单细胞转录组学在体内和体外鉴定软骨细胞分化动力学
IF 8.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
培养在体内复制的体外软骨细胞将有益于肌肉骨骼疾病建模和再生医学。尽管目前的方法已经取得了进展,但脱靶分化等挑战可能导致异质细胞状态。此外,缺乏与人类胚胎组织的比较妨碍了对体外细胞的详细评估。在这里,我们对胚胎长骨进行单细胞RNA测序(scRNA-seq),并将其与公开数据相结合,形成软骨内成骨图谱。我们用它来评估已发表的使用人类细胞系的体外软骨形成方案,发现每种细胞产生的差异性。我们将单核RNA测序(snRNA-seq)应用于我们的人类胚胎干细胞软骨形成方案,并与体内数据进行轨迹比对,以阐明体外脱靶分化。利用这些信息,我们抑制fox01,一种在胚胎成骨细胞和体外细胞中活跃的转录因子,以增加体外软骨细胞转录。这项工作为利用发育数据改善体外软骨形成提供了一个框架。本文章由计算机程序翻译,如有差异,请以英文原文为准。
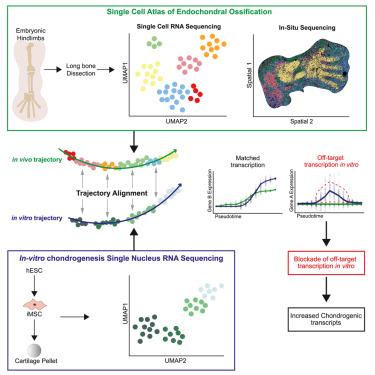
Single-cell transcriptomics identifies chondrocyte differentiation dynamics in vivo and in vitro
Developing in vitro chondrocytes that replicate in vivo development would benefit musculoskeletal disease modeling and regenerative medicine. Although current methodologies have made progress, challenges such as off-target differentiation can result in heterogeneous cell states. Furthermore, the lack of comparison with human embryonic tissue precludes detailed evaluation of in vitro cells. Here, we perform single-cell RNA sequencing (scRNA-seq) of embryonic long bones and combine this with public data to form an atlas of endochondral ossification. We use this to evaluate published in vitro chondrogenesis protocols that use human cell lines, finding variability in cells produced by each. We apply single-nuclear RNA sequencing (snRNA-seq) to our human embryonic stem cell chondrogenesis protocol and perform trajectory alignment with in vivo data to shed light on off-target differentiation in vitro. Using this information, we inhibit FOXO1, a transcription factor active in embryonic osteoblasts and in vitro cells, to increase chondrocyte transcripts in vitro. This work offers a framework for improving in vitro chondrogenesis using developmental data.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


