两个异源四倍体棉花种质的基因组组装揭示了体细胞胚胎发生机制,并实现了精确的基因组编辑
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
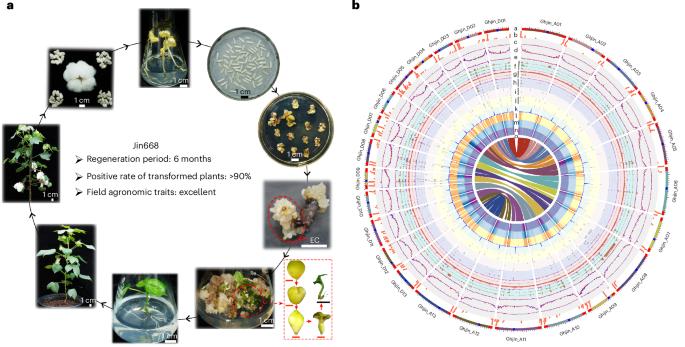
体细胞胚胎发生对植物基因工程至关重要,但棉花的潜在机制仍然知之甚少。在这里,我们提出了端粒到端粒的组装Jin668和高质量的组装YZ1,两个高度再生的异源四倍体棉花种质。Jin668基因组的完成可以表征玉米和Tekay逆转录转座子的着丝粒侵入的~30.1 Mb的着丝粒区域,一个包含25,190个拷贝的~8.1 Mb的5S rDNA阵列和一个包含8,131个拷贝的~75.1 Mb的主45S rDNA阵列。再生和顽固性基因型的比较分析揭示了在初始再生过程中的动态转录模式和染色质可及性。分层基因调控网络确定AGL15是再生的贡献者。此外,我们证明了遗传变异会影响sgRNA靶点,而Jin668基因组组装降低了基于crispr的基因组编辑中脱靶效应的风险。总之,完整的Jin668基因组揭示了基因组区域和棉花再生的复杂性,提高了基因组编辑的精度。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Genome assembly of two allotetraploid cotton germplasms reveals mechanisms of somatic embryogenesis and enables precise genome editing
Somatic embryogenesis is crucial for plant genetic engineering, yet the underlying mechanisms in cotton remain poorly understood. Here we present a telomere-to-telomere assembly of Jin668 and a high-quality assembly of YZ1, two highly regenerative allotetraploid cotton germplasms. The completion of the Jin668 genome enables characterization of ~30.1 Mb of centromeric regions invaded by centromeric retrotransposon of maize and Tekay retrotransposons, an ~8.1 Mb 5S rDNA array containing 25,190 copies and a ~75.1 Mb major 45S rDNA array with 8,131 copies. Comparative analyses of regenerative and recalcitrant genotypes reveal dynamic transcriptional patterns and chromatin accessibility during the initial regeneration process. A hierarchical gene regulatory network identifies AGL15 as a contributor to regeneration. Additionally, we demonstrate that genetic variation affects sgRNA target sites, while the Jin668 genome assembly reduces the risk of off-target effects in CRISPR-based genome editing. Together, the complete Jin668 genome reveals the complexity of genomic regions and cotton regeneration, and improves the precision of genome editing. Genome assemblies of two allotetraploid cotton germplasms, Jin668 and YZ1, reveal the regulatory mechanisms underlying somatic embryogenesis and plant regeneration, and provide potential for precise genome editing in cotton.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


