利用合理的β链插入策略和二硫锁来机械地操纵结构域交换的蛋白质结构
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
在畴交换中,连接两个任意结构域的畴交换结构的“铰环”区域发生了较大的结构改变。虽然以前的研究已经阐明了铰链区域在改变蛋白质寡聚状态中的作用,但我们的研究强调了如何精心操纵DS二聚体蛋白中的铰链环区域以产生显着改变的蛋白质结构,而无需改变蛋白质的寡聚状态。我们通过利用β链“之字形”构象的基本原理,说明了铰链区域中奇数与偶数氨基酸插入如何改变二级结构。这些微妙的变化导致了DS二聚体整体三维结构的可预测构象变化,同时使用二硫键交联策略降低了结构的域间灵活性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
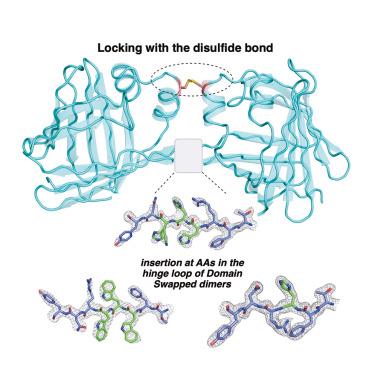
Exploiting a rational β-strand insertion strategy and disulfide locking to mechanically manipulate domain-swapped protein structures
In domain swapping, the “hinge loop” region of the domain-swapped (DS) structure that connects two structurally arbitrary domains undergoes a large structural alteration. While previous studies have shed light on the role of the hinge region in changing the oligomerization state of proteins, our study highlights how the hinge loop region in a DS dimer protein can be meticulously manipulated to generate significantly altered protein structures without the need to change the oligomeric state of the protein. We illustrate how an odd versus even number of amino acid insertions in the hinge region alters the secondary structure by exploiting the basic principle of the beta strand “zigzag” conformation. These subtle changes result in predictable conformational alterations in the overall 3D structure of DS dimers while reducing the interdomain flexibility of the structure using a disulfide bond cross-linking strategy.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


