单细胞和空间多组学研究了机械胁迫下杨树木质部的异质发育
IF 8.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
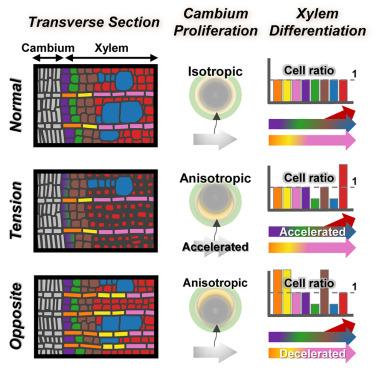
木质部是主要的结构支撑组织,在机械应力作用下形成具有丰富g层纤维的张力木材。尽管人们在一个多世纪前就认识到这一点,但三个关键的生物学问题仍然不清楚:(1)正常木材和张力木材中的纤维是否由于形态差异而不同?(2)张力木纤维是否来自不同的谱系?(3)控制张力木形成的关键基因有哪些?我们对正常、张力和对生木质部进行了单细胞RNA测序。正常木材和张力木材的纤维属于相同的细胞类型和谱系。空间转录组学和代谢组学进一步证实了张力木质部和对生木质部发育速度和细胞型比的差异。磷蛋白质组学显示了被子植物茎和根之间保守的机械传感机制。我们鉴定了一组参与张力木细胞命运转变的基因。关于细胞发育异质性的知识为优化生物质生产和生物能源产量提供了见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Single-cell and spatial multiomics identifies heterogeneous xylem development driven by mechanical stress in Populus
Xylem, the predominant tissue for structural support, forms tension wood with G-layer-rich fibers under mechanical stress. Despite being recognized over a century ago, three key biological questions remained unclear: (1) are fibers in normal and tension wood distinct cells due to morphological differences? (2) Do tension wood fibers arise from different lineages? (3) What are the key genes controlling tension wood formation? We conducted single-cell RNA sequencing on normal, tension and opposite xylem. Fibers in normal and tension wood belong to the same cell type and lineage. Differential developmental speed and cell-type ratio in tension and opposite xylem were further validated by spatial transcriptomics and metabolomics. Phosphoproteomics showed mechanical sensing mechanisms conserved between stems and roots across angiosperms. We identified a group of genes involved in the cell fate transition in tension wood. The knowledge on the heterogeneity of cell development offers insights into optimizing biomass production and bioenergy yield.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


