基于几何感知的低温电子层析图模板匹配
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
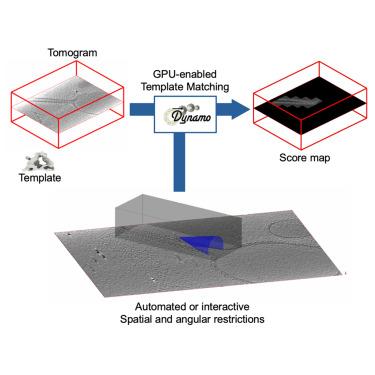
模板匹配作为冷冻电子断层扫描(cryo-ET)中体积自动分析的工具已有很长的历史。最近对该技术的理论和计算研究已经明确了使用细角度采样和高分辨率扫描来获得有意义的结果的重要性,从而强调了减轻该技术固有计算负担的方法的必要性。我们在Dynamo中提出了模型感知模板匹配的新模块,这是一个开源工具,允许在模板匹配计算的设置中集成可能可用的先验信息,在有利的情况下,由于扫描工作可以更有效地限制在相关的空间位置和动态定义的角度方向上,因此可以获得大量的计算收益。这种方法已经成功地在典型的断层扫描中产生的具有代表性的样品几何上进行了测试,模拟了分布在管状、膜状或泡状结构上的颗粒。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Geometry-aware template matching for cryo-electron tomograms in Dynamo
Template matching has a long history of serving as a tool for the automated analysis of volumes in cryo-electron tomography (cryo-ET). Recent theoretical and computational studies of the technique have pinpointed the importance of using fine angular samplings and high resolution scans to attain meaningful results, thus highlighting the necessity of approaches that alleviate the computational burden inherent to this technique. We present the new module for model-aware template matching in Dynamo, an open-source tool that allows the integration of possibly available a priori information in the set-up of a template matching computation, leading—in favorable cases—to large computational gains, as the scanning effort can be more efficiently restricted to relevant spatial positions and dynamically defined angular orientations. This approach has been successfully tested on a representative range of sample geometries typically arising in tomography, modeling particles distributed over tubular, membranous, or vesicular structures.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


