基于圆柱展开的螺旋参数精确估计
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
螺旋结构是丝状和管状大分子组装的基础,在结构支架和信号过程中起着至关重要的作用。这些组件的结构确定依赖于它们的螺旋参数的精确估计。虽然基于层线的方法被广泛使用,但由于其结构的灵活性和异质性,其应用往往具有挑战性。本文提出了一种基于圆柱展开的螺旋参数估计方法,称为螺旋简单(HELIS),并在同名软件包中实现。HELIS将螺旋结构视为滚动的二维(2D)晶体。低温电子层析成像(cryo-ET)显示结构的螺旋参数估计是由简化的二维往复式晶格测量得到的。HELIS还结合了辅助算法来跟踪和解开弯曲的细丝,确定相对极性,并执行原位螺旋重建。HELIS是一种适用于各种螺旋总成的高精度螺旋参数综合测定工具。本文章由计算机程序翻译,如有差异,请以英文原文为准。
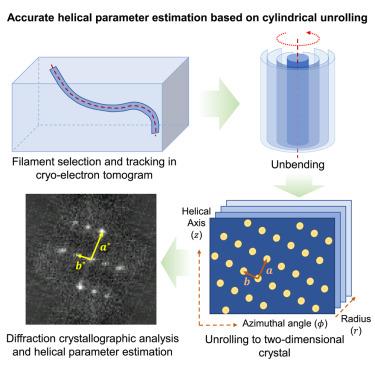
Accurate helical parameter estimation based on cylindrical unrolling
Helical structure is fundamental for filamentous and tubular macromolecular assemblies that play crucial roles in structural scaffolding and signaling processes. Structure determination of these assemblies relies on the precise estimation of their helical parameters. Although layer-line-based methods are widely used, their application is often challenging due to structural flexibility and heterogeneity. Herein, we report a method for helical parameter estimation based on cylindrical unrolling, called helix is simple (HELIS), which is implemented in a software package with the same name. HELIS treats helical structures as rolled two-dimensional (2D) crystals. Estimation of the helical parameters of structures visualized by cryo-electron tomography (cryo-ET) is derived from simplified 2D reciprocal-lattice measurements. HELIS also incorporates auxiliary algorithms to trace and unbend curved filaments, determine relative polarity, and perform in situ helical reconstruction. HELIS is a comprehensive helical structure determination tool with high accuracy for helical parameter estimation that is applicable to various helical assemblies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


