牛津纳米孔技术在非洲人类传染病检测和监测中的应用:范围综述
摘要
背景。基于纳米孔的测序由牛津纳米孔技术(ONT)提供快速,经济高效和便携式测序。作为一项新兴技术,ONT必须在资源丰富和资源贫乏的环境中进行有效性和实际应用评估。这篇范围综述(SR)旨在(1)描述纳米孔技术如何在非洲用于人类传染病的监测和诊断,(2)描述纳米孔技术如何帮助非洲实时检测传染性病原体,以及(3)确定利用纳米孔技术在非洲研究传染病的挑战和机遇。方法。该SR遵循了乔安娜布里格斯研究所的SR评审手册框架。纳入了2008年1月1日至2024年4月30日发表的在非洲采集的人类标本上使用ONT并针对≥1种微生物制剂的英语研究。在Embase、Medline、PubMed、CINAHL和Cochrane图书馆进行检索。在数据收集之前,该方案已在开放科学框架Bastug等人(非洲传染病监测和诊断的纳米孔测序:2024年范围审查)上公开提供。两名独立审稿人筛选了使用covid - ence的研究,并使用定制的REDCap仪器提取数据。在Microsoft Excel中进行描述性统计和数据可视化。结果。共确定了1262项研究,其中93项(8%)进行了全文综述。便携式MinION Mk1B是最常见的ONT设备(65%的研究)。88项研究分析了来自一个非洲国家的标本。其中,45%在同一国家测序,7%在不同的非洲国家测序,11%在非非洲国家测序,而32%没有指定位置。标本类型包括直接患者标本(62%)和培养分离株(35%),或两者的组合。血液、血清或血浆是最常见的(35%),其次是鼻或口咽标本(27%)。44项研究在活跃的传染病暴发期间使用了ONT,其中25项研究研究了严重急性呼吸综合征冠状病毒2 (SARS-CoV-2)。72项研究将ONT用于感染性病原体或抗生素耐药基因的基因组监测,一项研究将ONT用于直接临床应用。在46%的研究中,与非洲有关联的作者被列为第一、中间和最后作者,15%的研究由完全与非洲有关联的团队发表。10项研究公布了工作流程时间线信息,5项研究公布了每个样本的成本。结论。通过个别实现这些目标的少数研究表明,ONT可以使非洲国家能够及时和负担得起测序。大多数研究将ONT用于病原体或抗菌素耐药基因的基因组监测,而只有一项研究直接将ONT用于实时临床应用。少数研究描述了标本采集和测序结果之间的短间隔,支持临床应用的可能性。有必要改进ONT方法的报告,包括管道时间表、成本、条形码的使用、流动池模型和阴性对照的使用。提供这些细节的出版物将提高可重复性,并支持在资源匮乏的环境中开展利用ONT诊断和监测传染病的新研究。

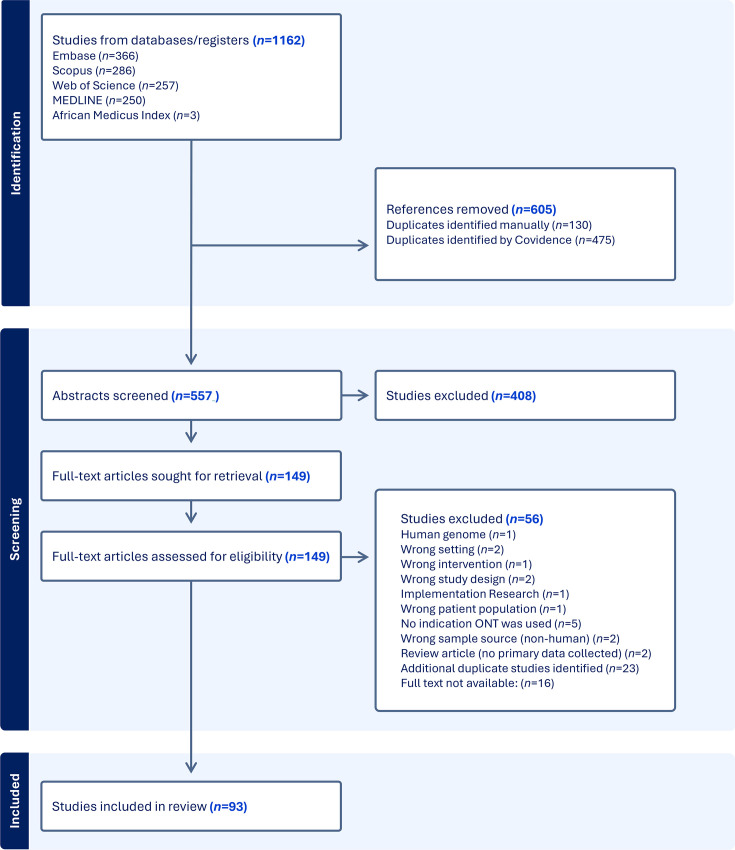

Background. Nanopore-based sequencing by Oxford Nanopore Technologies (ONT) offers rapid, cost-effective and portable sequencing. As an emerging technology, ONT must be evaluated for efficacy and practical application in both high- and low-resource settings. This scoping review (SR) aimed to (1) describe how nanopore technology is used in Africa for surveillance and diagnosis of human infectious diseases, (2) describe how nanopore technology aids in the real-time detection of infectious pathogens in Africa and (3) identify challenges and opportunities for utilizing nanopore technology in Africa to study infectious diseases. Methods. This SR followed the Joanna Briggs Institute Reviewer's Manual framework for SRs. English language studies published from 1 January 2008 to 30 April 2024 that used ONT on human specimens collected in Africa and targeted ≥1 microbial agent were included. Searches were performed in Embase, Medline, PubMed, CINAHL and the Cochrane Library. The protocol was publicly available on the Open Science Framework Bastug et al. (Nanopore Sequencing for Infectious Diseases Surveillance and Diagnostics in Africa: a Scoping Review 2024) prior to data collection. Two independent reviewers screened studies using Covidence, and data was extracted using a custom REDCap instrument. Descriptive statistics and data visualization were performed in Microsoft Excel. Results. One thousand one hundred sixty-two studies were identified and 93 (8%) underwent full-text review. The portable MinION Mk1B was the most common ONT device (65% of studies). Eighty-eight studies analysed specimens from a single African country. Of these, 45% were sequenced in the same country, 7% in a different African country and 11% in a non-African country, while 32% did not specify the location. Specimen types included direct patient specimens (62%) and cultured isolates (35%), or a combination of both. Blood, serum or plasma was most common (35%), followed by naso- or oropharyngeal specimens (27%). Forty-four studies used ONT during an active infectious disease outbreak, 25 of which studied severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Seventy-two studies used ONT for genomic surveillance of infectious pathogens or antibiotic resistance genes, and one study used ONT for a direct clinical application. African-affiliated authors were included as first, middle and last authors in 46% of studies, and 15% were published by entirely African-affiliated teams. Ten studies published information on workflow timeline, and five studies published the per-specimen cost. Conclusions. ONT can enable timely and affordable sequencing in African countries as demonstrated through a small number of studies that accomplished these goals individually. Most studies used ONT for genomic surveillance of pathogens or antimicrobial resistance genes, while only one study used ONT directly for a real-time clinical application. A small number of studies described a short interval between specimen collection and sequence result, supporting that clinical applications are possible. There is a need for improved reporting of ONT methodology including pipeline timelines, cost, use of barcoding, flow cell models and the use of negative controls. Publications that provide these details will enhance reproducibility and support the development of new studies using ONT for the diagnosis and surveillance of infectious diseases in low-resource settings.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


