三家医院联合携带blaNDM-1和mcr-9的弗氏柠檬酸杆菌和阴沟肠杆菌复合体的鉴定
IF 3.2
3区 医学
Q2 INFECTIOUS DISEASES
引用次数: 0
摘要
目的:抗菌素耐药性(AMR)是一个全球性的健康问题,与抗菌素使用和随后出现的耐药生物有关,包括产碳青霉烯酶肠杆菌(CPE)。CPE分离株同时携带blaNDM-1和mcr-9.1在国际上报道甚少。在一个地区的三家医院内发现了20种这样的分离株,其中16种属于同一种,表明可能在卫生保健机构内部和机构之间传播。方法:采用MALDI-ToF ms对20株菌株进行伪匿名鉴定,采用纸片扩散法进行药敏试验,采用UMIC系统对黏菌素进行最小抑制浓度测定。利用Illumina MiSeq平台进行短读测序,基因组分析鉴定了抗菌素耐药基因、毒力因子和质粒contigs。利用Kraken2对草稿基因组进行生物信息学分类。结果:该收集包括霍氏肠杆菌(n=16)、弗氏Citrobacter freundii (n=3)和阴沟肠杆菌(n=1),它们来自常规筛查(n=13)或医疗保健相关环境站点(n=7)收集的患者直肠棉签。该菌株包括4种不同的ST型,一种基于未分配ST组的质粒分析鉴定出17种质粒复制子类型。所有分离株均检测到IncHI2A、IncHI2和pKPC-CAV1321_1。相关blaNDM-1和mcr-9.1基因在医院的传播可能是通过质粒介导的转移而不是荷氏肠杆菌的传播。结论:该研究是迄今为止欧洲首次记录的blaNDM-1/mcr-9.1共存病例。它强调了抗菌素耐药性对公共卫生造成的日益严重的威胁,并强调了基因组监测和临床筛查的重要性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
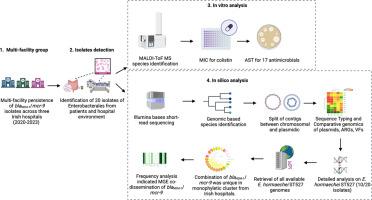
Characterisation of Citrobacter freundii and Enterobacter cloacae complex isolates co-carrying blaNDM-1 and mcr-9 from three hospitals
Objectives
Antimicrobial resistance (AMR) is a global health concern related to antimicrobial use and the subsequent emergence of resistant organisms, including carbapenemase-producing Enterobacterales (CPE). CPE isolate co-carrying blaNDM-1 and mcr-9.1 have been scarcely reported internationally. The identification of 20 such isolates, including 16 of one species, within a group of three hospitals in one region indicated potential dissemination within and between healthcare facilities.
Methods
Twenty isolates were pseudo-anonymised and identified via MALDI-ToF MS. Antimicrobial susceptibility testing was performed by disc diffusion, and Minimal Inhibition Concentration for colistin was carried out using the UMIC system. Short-read sequencing was conducted using the Illumina MiSeq platform, and genomic analysis identified antimicrobial resistance genes, virulence factors and plasmid contigs. Taxonomic classification of draft genomes was bioinformatically assessed using Kraken2.
Results
This collection comprised of Enterobacter hormaechei (n = 16), Citrobacter freundii (n = 3) and Enterobacter cloacae (n = 1) sourced from patient rectal swabs collected during routine screening (n = 13) or from healthcare-associated environmental sites (n = 7). The E. hormaechei isolates included four different ST types with one unassigned ST. Contig-based plasmid analysis identified 17 plasmid replicon types among the isolates. IncHI2A, IncHI2, and pKPC![]() CAV1321_1 were detected in all isolates. Linked blaNDM-1 and mcr-9.1 gene spread in hospitals likely occurred via plasmid-mediated transfer rather than spread of E. hormaechei.
CAV1321_1 were detected in all isolates. Linked blaNDM-1 and mcr-9.1 gene spread in hospitals likely occurred via plasmid-mediated transfer rather than spread of E. hormaechei.
Conclusions
This study represents the first documented instance of blaNDM-1/mcr-9.1 co-occurrence in Europe to date. It highlights the increasing public health threat posed by antimicrobial resistance and underscores the importance of genomic surveillance and clinical screening.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of global antimicrobial resistance
INFECTIOUS DISEASES-PHARMACOLOGY & PHARMACY
CiteScore
8.70
自引率
2.20%
发文量
285
审稿时长
34 weeks
期刊介绍:
The Journal of Global Antimicrobial Resistance (JGAR) is a quarterly online journal run by an international Editorial Board that focuses on the global spread of antibiotic-resistant microbes.
JGAR is a dedicated journal for all professionals working in research, health care, the environment and animal infection control, aiming to track the resistance threat worldwide and provides a single voice devoted to antimicrobial resistance (AMR).
Featuring peer-reviewed and up to date research articles, reviews, short notes and hot topics JGAR covers the key topics related to antibacterial, antiviral, antifungal and antiparasitic resistance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


