纳米孔靶向测序揭示BRCA1中隐藏的SVA反转录转座子插入导致遗传性乳腺癌和卵巢癌。
IF 2.5
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
摘要
在日本,种系BRCA1/2基因检测被广泛用于遗传性乳腺癌和卵巢癌综合征(HBOC)的诊断。然而,不确定的结果有时使临床管理复杂化。在本研究中,我们通过应用纳米孔自适应采样和Flongle基因组扩增子测序,通过靶向长读测序(LRS),在一个先证者及其母亲的BRCA1基因中发现了一个内含子sin - vntr - alu (SVA)插入,这两个先证者的BRCA1/2基因检测结果都不确定。我们进一步利用TaqMan探针和Flongle cDNA扩增子测序的cDNA定量PCR证实了剪接畸变。我们的研究结果强调,除了传统的BRCA1/2基因检测外,在某些情况下,使用靶向LRS进行结构变异分析对于准确诊断HBOC是必不可少的。此外,Flongle扩增子测序被证明对传统PCR和Sanger测序难以测序的区域有效,特别是重复和富含gc的区域,如反转录转座子。本文章由计算机程序翻译,如有差异,请以英文原文为准。
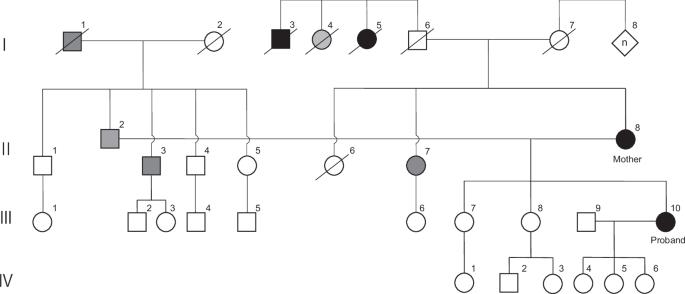
Hidden SVA retrotransposon insertion in BRCA1 revealed by nanopore targeted sequencing causes hereditary breast and ovarian cancer
In Japan, germline BRCA1/2 genetic testing is extensively used for the diagnosis of hereditary breast and ovarian cancer syndrome (HBOC). However, inconclusive results sometimes complicate clinical management. In this study, we identified an intronic SINE-VNTR-Alu (SVA) insertion in BRCA1 of a proband and her mother, both of whom had inconclusive conventional BRCA1/2 genetic test results, by targeted long-read sequencing (LRS) through the application of nanopore adaptive sampling and Flongle genome amplicon sequencing. We further confirmed splicing aberrations using cDNA quantitative PCR with TaqMan probes and Flongle cDNA amplicon sequencing. Our findings highlighted that, in addition to conventional BRCA1/2 genetic testing, structural variation analysis using targeted LRS is indispensable for the accurate diagnosis of HBOC in certain cases. Furthermore, Flongle amplicon sequencing was demonstrated to be effective for sequencing regions refractory to conventional PCR and Sanger sequencing, particularly repetitive and GC-rich regions, such as retrotransposons.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Human Genetics
生物-遗传学
CiteScore
7.20
自引率
0.00%
发文量
101
审稿时长
4-8 weeks
期刊介绍:
The Journal of Human Genetics is an international journal publishing articles on human genetics, including medical genetics and human genome analysis. It covers all aspects of human genetics, including molecular genetics, clinical genetics, behavioral genetics, immunogenetics, pharmacogenomics, population genetics, functional genomics, epigenetics, genetic counseling and gene therapy.
Articles on the following areas are especially welcome: genetic factors of monogenic and complex disorders, genome-wide association studies, genetic epidemiology, cancer genetics, personal genomics, genotype-phenotype relationships and genome diversity.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


