单细胞空间转录组学揭示了AML中免疫治疗驱动的骨髓生态位重塑
IF 12.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
鉴于同种异体造血干细胞移植在难治性或复发性急性髓性白血病(AML)中观察到的移植物抗白血病效应,免疫疗法已经在非移植环境中进行了探索。我们应用多组学方法来检测接受派姆单抗和地西他滨治疗的AML患者骨髓相互作用。使用广泛训练的核和膜分割模型,我们实现了精确的转录本分配和基于深度学习的图像分析。为了解决读取深度的限制,我们将来自同一样本的单细胞RNA测序与单细胞空间转录组学相结合。在边缘水平定量细胞-细胞距离可以更准确地分析肿瘤微环境,揭示白血病细胞附近的全局和局部免疫细胞富集,这可能与临床反应有关。此外,配体受体分析表明,在免疫治疗后,白血病和免疫细胞之间的特定信号通路可能发生改变。这些发现为AML的免疫相互作用提供了见解,并可能为治疗策略提供信息。本文章由计算机程序翻译,如有差异,请以英文原文为准。
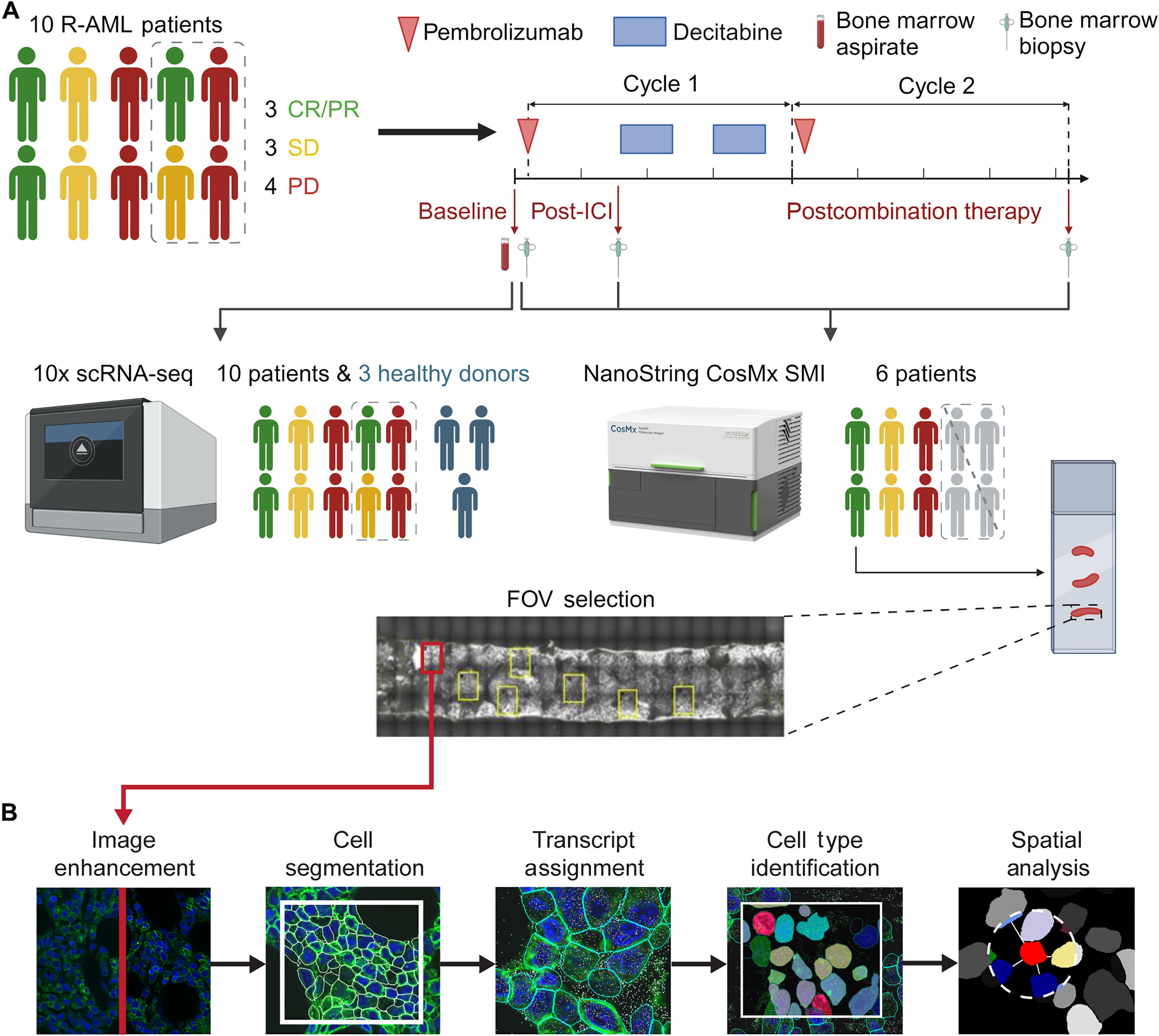
Single-cell spatial transcriptomics reveals immunotherapy-driven bone marrow niche remodeling in AML
Given the graft-versus-leukemia effect observed with allogeneic hematopoietic stem cell transplantation in refractory or relapsed acute myeloid leukemia (AML), immunotherapies have been explored in nontransplant settings. We applied a multiomic approach to examine bone marrow interactions in patients with AML treated with pembrolizumab and decitabine. Using extensively trained nuclear and membrane segmentation models, we achieved precise transcript assignment and deep learning–based image analysis. To address read-depth limitations, we integrated single-cell RNA sequencing with single-cell spatial transcriptomics from the same sample. Quantifying cell-cell distances at the edge level enabled more accurate tumor microenvironment analysis, revealing global and local immune cell enrichment near leukemia cells postpembrolizumab treatment, potentially linked to clinical response. Furthermore, ligand-receptor analysis indicated potential alterations in specific signaling pathways between leukemia and immune cells following immunotherapy treatment. These findings provide insights into immune interactions in AML and may inform therapeutic strategies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Science Advances
综合性期刊-综合性期刊
CiteScore
21.40
自引率
1.50%
发文量
1937
审稿时长
29 weeks
期刊介绍:
Science Advances, an open-access journal by AAAS, publishes impactful research in diverse scientific areas. It aims for fair, fast, and expert peer review, providing freely accessible research to readers. Led by distinguished scientists, the journal supports AAAS's mission by extending Science magazine's capacity to identify and promote significant advances. Evolving digital publishing technologies play a crucial role in advancing AAAS's global mission for science communication and benefitting humankind.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


