西班牙全国范围内铜绿假单胞菌头孢醚抗微生物药物敏感性和耐药性基因组学调查。
IF 4.6
2区 医学
Q1 INFECTIOUS DISEASES
International Journal of Antimicrobial Agents
Pub Date : 2025-07-01
DOI:10.1016/j.ijantimicag.2025.107563
引用次数: 0
摘要
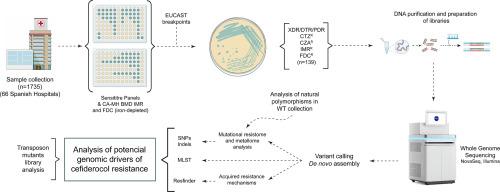
背景:Cefiderocol是一种新的铁载体-头孢菌素,对铜绿假单胞菌(P. aeruginosa)具有强效活性,包括广泛耐药(XDR)或难以治疗耐药(DTR)表型的分离株。然而,对潜在的耐药机制的了解仍然有限。目的:分析头孢地罗在西班牙全国范围内的活性,确定其对大多数相关耐药机制的稳定性,并通过全基因组测序(WGS)分析潜在的耐药驱动因素。方法:采用微量肉汤稀释法测定西班牙66家医院1735株Cefiderocol的mic,并与一组13种抗假单胞菌药物进行比较。所有XDR/DTR菌株以及对头孢氧杂酮/他唑巴坦、头孢他啶/阿维巴坦、亚胺培南/relebactam或头孢地罗耐药的菌株进行WGS (NovaSeq, Illumina)和生物信息学分析,包括开发用于评估获得性耐药决定因素、突变抗性组和铁摄取基因的特定管道。结果:头孢地罗对铜绿假单胞菌的敏感性最高(99.7%),且活性保守(结论:头孢地罗对铜绿假单胞菌具有较强的抗药活性,但应在表型和基因组水平上积极监测耐药菌株的出现。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Spanish nationwide survey of Pseudomonas aeruginosa cefiderocol antimicrobial susceptibility and resistance genomics
Objective
Cefiderocol is a new siderophore-cephalosporin showing potent activity against Pseudomonas aeruginosa, including isolates showing extensively drug-resistant (XDR) or difficult-to-treat resistant (DTR) phenotypes. However, there is still a limited understanding of the potential resistance mechanisms. The objective of this study was to analyse the activity of cefiderocol in a nationwide Spanish survey, determine its stability against most relevant resistance mechanisms, and analyse potential drivers of resistance through whole-genome sequencing.
Methods
Cefiderocol MICs were determined by broth microdilution in cation-adjusted iron-depleted Müller-Hinton broth for 1735 isolates from 66 Spanish hospitals and compared with those of a panel of 13 antipseudomonal agents. All XDR/DTR strains and those resistant to ceftolozane/tazobactam, ceftazidime/avibactam, imipenem/relebactam or cefiderocol were subjected to whole-genome sequencing (NovaSeq, Illumina, San Diego, California, US) and bioinformatic analysis, including specific pipelines developed for the assessment of acquired resistance determinants, the mutational resistome and iron-uptake genes.
Results
Cefiderocol showed the highest percentage of susceptibility (99.7%) and conserved activity (<5–10% resistance) against XDR/DTR strains and those producing carbapenemases. Only five resistant isolates were detected and the complex resistome included acquired β-lactamases or AmpC mutations along with mutations in iron-uptake systems and AmpC and/or efflux pump regulators. Novel drivers of resistance such as piuE were identified. Moreover, the production of acquired oxacillinases and mutations in ampC, mexR or piuA were statistically associated with increased MICs, whereas mutations in gyrA, parC or pvdS were associated with increased susceptibility.
Conclusion
While cefiderocol shows potent activity against P. aeruginosa, active surveillance at the phenotypic and genomic level for the emergence of resistant strains should be implemented.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
21.60
自引率
0.90%
发文量
176
审稿时长
36 days
期刊介绍:
The International Journal of Antimicrobial Agents is a peer-reviewed publication offering comprehensive and current reference information on the physical, pharmacological, in vitro, and clinical properties of individual antimicrobial agents, covering antiviral, antiparasitic, antibacterial, and antifungal agents. The journal not only communicates new trends and developments through authoritative review articles but also addresses the critical issue of antimicrobial resistance, both in hospital and community settings. Published content includes solicited reviews by leading experts and high-quality original research papers in the specified fields.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


