多组学整合揭示乳腺癌新辅助治疗反应的亚型特异性预测因子
IF 12.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
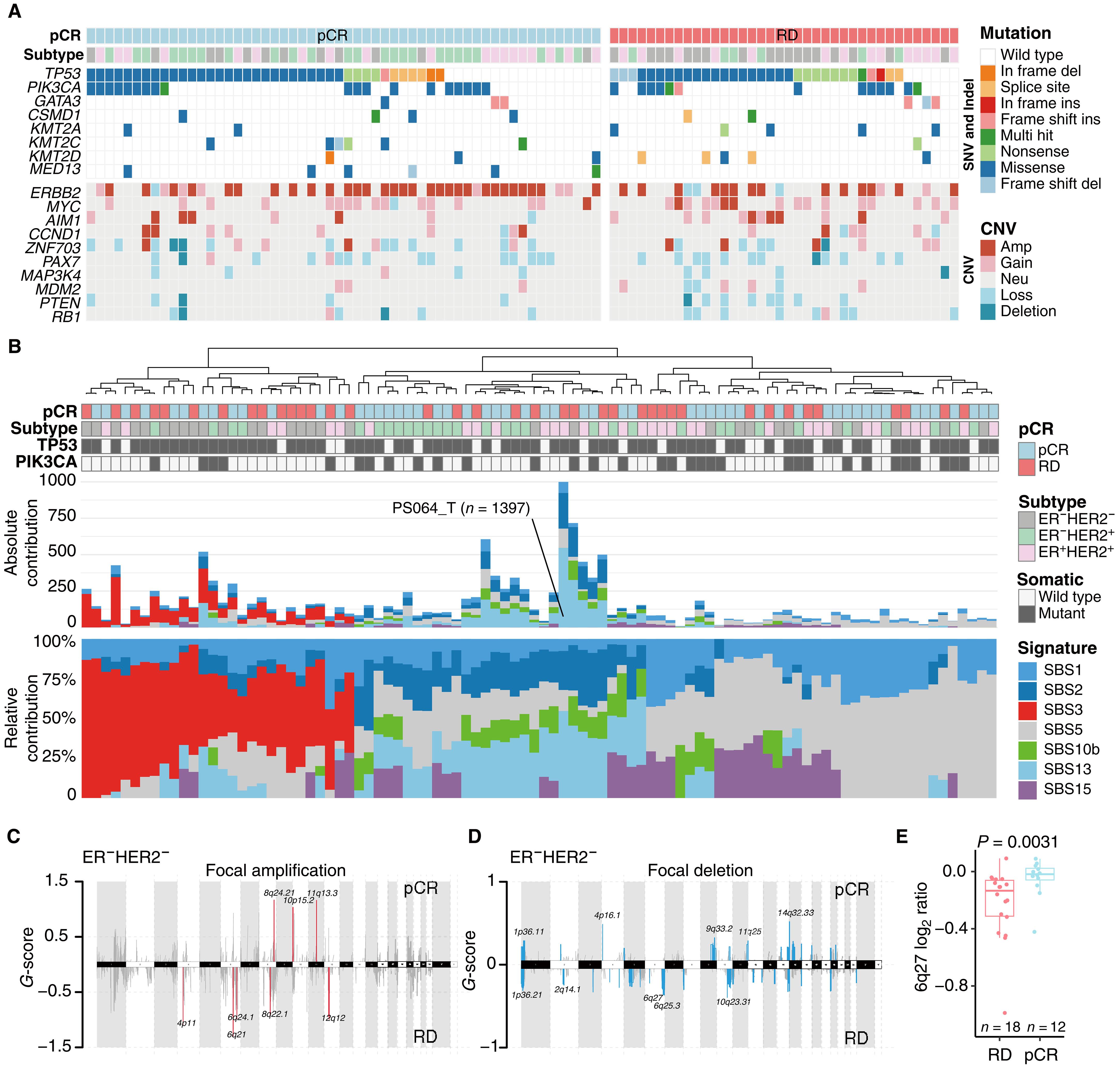
新辅助治疗已广泛应用于乳腺癌,但治疗反应因人而异。我们对149名中国乳腺癌患者的肿瘤样本进行了ER - HER2+、ER+HER2+和ER - HER2 -亚型的多组学分析,将结果分类为病理完全缓解(pCR;n = 81)或残留病变(RD;n = 68)。我们确定了每种亚型中与pCR相关的不同分子特征:ER - HER2 - pCR患者细胞增殖升高,ER - HER2 - RD患者CDKN2A甲基化升高,ER - HER2+ RD患者KIT甲基化升高,ER+HER2+ RD患者MAP4K1超甲基化。这些发现随后在独立数据集中得到验证。通过整合临床和多组学数据,我们开发了MOPCR,这是一种亚型特异性机器学习模型,在预测治疗反应方面优于单组学方法。MOPCR显示了跨队列的潜在普遍性,并提供了具有更高pCR概率的患者亚组的初步分层,为精确的癌症管理提供了有价值的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Multiomic integration reveals subtype-specific predictors of neoadjuvant treatment response in breast cancer
Neoadjuvant therapy has been widely used in breast cancer, but treatment response varies among individuals. We conducted multiomic profiling on tumor samples from 149 Chinese patients with breast cancer across ER−HER2+, ER+HER2+, and ER−HER2− subtypes, categorizing outcomes as pathologic complete response (pCR; n = 81) or residual disease (RD; n = 68). We identified distinct molecular features linked to pCR in each subtype: elevated cell proliferation in patients with ER−HER2− pCR, higher CDKN2A methylation in patients with ER−HER2− RD, increased KIT methylation in patients with ER−HER2+ RD, and MAP4K1 hypermethylation in patients with ER+HER2+ RD. These findings were subsequently validated in independent datasets. By integrating clinical and multiomic data, we developed MOPCR, a subtype-specific machine learning model that outperformed single-omic approaches in predicting treatment response. MOPCR demonstrated potential generalizability across cohorts and provided preliminary stratification of patient subgroups with higher pCR probability, offering valuable insights for precision cancer management.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Science Advances
综合性期刊-综合性期刊
CiteScore
21.40
自引率
1.50%
发文量
1937
审稿时长
29 weeks
期刊介绍:
Science Advances, an open-access journal by AAAS, publishes impactful research in diverse scientific areas. It aims for fair, fast, and expert peer review, providing freely accessible research to readers. Led by distinguished scientists, the journal supports AAAS's mission by extending Science magazine's capacity to identify and promote significant advances. Evolving digital publishing technologies play a crucial role in advancing AAAS's global mission for science communication and benefitting humankind.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


