研究早期人类胚胎外胚层细胞特化和分化的动态信号通路。
IF 2.1
3区 生物学
Q2 DEVELOPMENTAL BIOLOGY
引用次数: 0
摘要
人类胚胎外胚层对早期发育至关重要,最终形成所有体细胞和生殖细胞谱系。尽管对胚胎发生的了解有所进展,但在外胚层发育过程中调节细胞间通讯的机制仍然不完全清楚。在这里,我们分析了人类胚胎着床前、着床后、原肠胚形成前和早期原肠胚形成阶段的单细胞RNA测序数据,以研究胚胎外组织的信号如何影响外胚层(EPI)的发育。我们的数据驱动分析表明,多种信号通路,包括BMP, WNT, FGF, LIF和TGFβ,可能参与第一和第二谱系分离,羊膜上皮(AME)细胞发育和原始条纹(PS)形成。我们进一步提出内细胞团(ICM)和EPI可以作为信号中枢,协调关键的细胞间通讯事件。基于RNA表达模式,胚胎外组织,如下胚层、滋养层(TrB)和胚胎外中胚层(ExM)似乎分泌关键信号(BMP4、SELL、WNTs和PTN),这些信号可能调节EPI细胞极化、EPI- ame转分化和PS发育。值得注意的是,BMP4的表达可能遵循一个动态模式,从早期着床的内脏/顶叶内胚层(VE/YE)细胞过渡到原肠胚形成前的ExM细胞,最终过渡到CS7晚期中胚层细胞。总的来说,这些发现提供了由胚胎外组织介导的假定信号事件的全面概述,并强调了它们在人类胚胎中协调外胚层发育和早期谱系决定中的潜在作用,同时强调了进一步的蛋白质水平和功能验证的必要性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
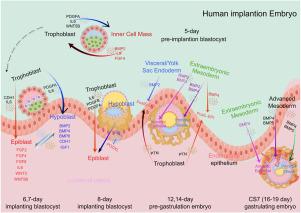
Investigating the dynamic signaling pathways involved in the specialization and differentiation of epiblast cells in early human embryos
The human embryonic epiblast is essential for early development, ultimately giving rise to all somatic and germ cell lineages. Despite advances in understanding embryogenesis, the mechanisms regulating intercellular communication during epiblast specification remain incompletely understood. Here, we analyzed single-cell RNA sequencing data spanning pre-implantation, post-implantation, pre-gastrulation, and early-gastrulation stages of human embryos to investigate how signals from extra-embryonic tissues influence epiblast (EPI) development. Our data-driven analysis indicates that multiple signaling pathways, including BMP, WNT, FGF, LIF, and TGF, may be involved in the first and second lineage separations, amniotic epithelial (AME) cell development, and primitive streak (PS) formation. We further propose that the inner cell mass (ICM) and EPI could function as signaling hubs, coordinating critical intercellular communication events. Based on RNA expression patterns, extra-embryonic tissues such as the hypoblast, trophoblast (TrB), and extra-embryonic mesoderm (ExM) appear to secrete key signals (BMP4, SELL, WNTs, and PTN) that potentially regulate EPI cell polarization, EPI-AME transdifferentiation, and PS development. Notably, BMP4 expression may follow a dynamic pattern, transitioning from early implantation visceral/parietal endoderm (VE/YE) cells to pre-gastrulation ExM cells and ultimately to CS7 advanced mesoderm cells. Overall, these findings provide a comprehensive overview of putative signaling events mediated by extra-embryonic tissues and underscore their potential roles in orchestrating epiblast development and early lineage decisions in the human embryo, while highlighting the need for further protein-level and functional validation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Developmental biology
生物-发育生物学
CiteScore
5.30
自引率
3.70%
发文量
182
审稿时长
1.5 months
期刊介绍:
Developmental Biology (DB) publishes original research on mechanisms of development, differentiation, and growth in animals and plants at the molecular, cellular, genetic and evolutionary levels. Areas of particular emphasis include transcriptional control mechanisms, embryonic patterning, cell-cell interactions, growth factors and signal transduction, and regulatory hierarchies in developing plants and animals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


