基于区域的U-nets用于快速、准确和可扩展的脑深部分割:在帕金森综合征中的应用
IF 3.6
2区 医学
Q2 NEUROIMAGING
引用次数: 0
摘要
与年龄相关的神经退行性疾病往往发展为痴呆症,由于这些疾病早期症状微妙且重叠,因此早期诊断构成了重大的临床挑战。自动MRI分割对于早期检测非常重要,因为它提供了一致的测量结果,并且能够检测到大脑中细微的结构变化。人工分割对于大数据集或临床使用是不切实际的。深度学习方法提供快速处理,然而,在处理大型数据集时,它们经常遇到图形处理单元(GPU)内存限制。在这里,我们介绍了一种基于深度学习的方法,使用基于区域的U-nets,专门设计用于分割与帕金森综合征相关的12个深层脑结构。通过将大脑图像划分为脑干、脑室系统和纹状体周围的目标区域,我们的方法优化了GPU的使用,显著减少了训练时间,同时保持了较高的准确性。在三个数据集上验证了所提出的方法,其中包括660个受试者的临床数据集,包括健康对照和各种运动障碍患者,我们证明了鲁棒性和实际适用性,可以区分不同的疾病。与现有的分割方法相比,该方法具有更好的分割性能,平均骰子相似系数(DSC)为0.90,95%豪斯多夫距离(HD95)为1.35 mm,平均对称表面距离(ASSD)为0.45 mm,显示了其分割的准确性和鲁棒性。此外,我们的方法优于这些方法,将训练时间从几天减少到几个小时,同时提供每个主题不到一秒的处理时间。源代码和训练模型将在GitHub上公开提供。本文章由计算机程序翻译,如有差异,请以英文原文为准。
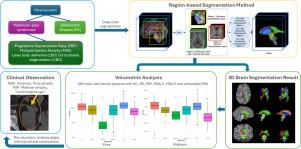
Region-based U-nets for fast, accurate, and scalable deep brain segmentation: Application to Parkinson Plus Syndromes
The early diagnosis of age-related neurodegenerative diseases, which often progress to dementia, poses significant clinical challenges due to subtle and overlapping symptoms of these diseases at early stage. Automated MRI segmentation is important for early detection, as it offers consistent measurements and the ability to detect subtle structural changes in the brain. Manual segmentation is impractical for large datasets or clinical use. Deep learning approaches provide fast processing, however, they often encounter graphics processing unit (GPU) memory constraints when handling large datasets. Here we introduce a deep learning-based approach using region-based U-nets specifically designed to segment 12 deep-brain structures relevant to Parkinson Plus Syndromes. By dividing the brain image into targeted regions around the brainstem, ventricular system, and striatum, our method optimizes GPU usage and significantly reduces training times, while maintaining high accuracy. Validating the proposed method on three datasets, including a 660-subject clinical dataset comprising both healthy controls and patients with various movement disorders, we demonstrate robustness and practical applicability in separating different diseases. The method achieves superior segmentation performance compared to state-of-the-art methods, with a mean Dice Similarity Coefficient (DSC) of 0.90, a 95% Hausdorff Distance (HD95) of 1.35 mm, and an Average Symmetric Surface Distance (ASSD) of 0.45 mm, showcasing its segmentation accuracy and robustness. Furthermore, our method outperforms these methods by reducing training time from several days to a few hours while providing a processing time of less than a second per subject. The source code and trained model will be made publicly available on GitHub.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Neuroimage-Clinical
NEUROIMAGING-
CiteScore
7.50
自引率
4.80%
发文量
368
审稿时长
52 days
期刊介绍:
NeuroImage: Clinical, a journal of diseases, disorders and syndromes involving the Nervous System, provides a vehicle for communicating important advances in the study of abnormal structure-function relationships of the human nervous system based on imaging.
The focus of NeuroImage: Clinical is on defining changes to the brain associated with primary neurologic and psychiatric diseases and disorders of the nervous system as well as behavioral syndromes and developmental conditions. The main criterion for judging papers is the extent of scientific advancement in the understanding of the pathophysiologic mechanisms of diseases and disorders, in identification of functional models that link clinical signs and symptoms with brain function and in the creation of image based tools applicable to a broad range of clinical needs including diagnosis, monitoring and tracking of illness, predicting therapeutic response and development of new treatments. Papers dealing with structure and function in animal models will also be considered if they reveal mechanisms that can be readily translated to human conditions.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


