分裂转录因子增强子在果蝇R7光感受器规范中的作用
IF 2.1
3区 生物学
Q2 DEVELOPMENTAL BIOLOGY
引用次数: 0
摘要
当细胞同时接收多个发育信号时,这些信号触发的细胞内转导通路同时活跃。那么,细胞如何解码包含在这些多种活跃途径中的信息,从而得出精确的发育指令呢?果蝇R7光感受器的描述是研究这类问题的经典模型系统。R7的命运是由Notch (N)和受体酪氨酸激酶(RTK)信号通路的联合作用决定的。这两种通路在一个整合的机制中交叉交流,也相互独立地提供信息。总的来说,这些信息为R7的命运提供了一个明确的指示。我们的目标是理解这些机制。在这里,我们研究了N活性如何在与RTK通路的信息整合过程中抑制叶足类基因的转录,以及它如何在R7命运所需的独立机制中抑制7 -up基因的表达。我们描述了N活性如何实现这些转录抑制,并确定分裂转录因子的增强子作为这些作用的介质。本文章由计算机程序翻译,如有差异,请以英文原文为准。
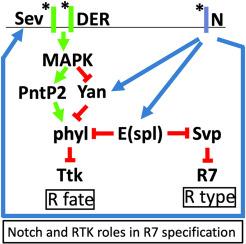
Roles played by Enhancer of split transcription factors in Drosophila R7 photoreceptor specification
When a cell receives multiple developmental signals simultaneously, the intracellular transduction pathways triggered by those signals are coincidentally active. How then, do the cells decode the information contained within those multiple active pathways to derive a precise developmental directive? The specification of the Drosophila R7 photoreceptor is a classic model system for investigating such questions. The R7 fate is specified by the combined actions of the Notch (N) and receptor tyrosine kinase (RTK) signaling pathways. The two pathways cross-communicate in an integrative mechanism and also supply information independently of each other. Collectively, this information is summed to provide an unambiguous directive for the R7 fate. Our goal is to understand these mechanisms. Here, we examine how N activity represses transcription of the phyllopod gene in the process of information integration with the RTK pathway, and how it represses expression of the seven-up gene in an independent mechanism needed for R7 fate. We describe how N activity achieves these transcriptional repressions and identify Enhancer of Split transcription factors as the mediators of its functions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Developmental biology
生物-发育生物学
CiteScore
5.30
自引率
3.70%
发文量
182
审稿时长
1.5 months
期刊介绍:
Developmental Biology (DB) publishes original research on mechanisms of development, differentiation, and growth in animals and plants at the molecular, cellular, genetic and evolutionary levels. Areas of particular emphasis include transcriptional control mechanisms, embryonic patterning, cell-cell interactions, growth factors and signal transduction, and regulatory hierarchies in developing plants and animals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


