蜻蜓和豆娘端粒重复序列(TTGGG)的获取和重复改变。
IF 3.7
2区 农林科学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
大多数真核生物的染色体末端由简单的端粒重复组成。对于节肢动物,TTAGG五核苷酸重复序列(TTAGG)n被认为是祖先的端粒重复序列。然而,在蜻蜓目,包括蜻蜓和豆豆蝇的最早的昆虫分化类群中,(TTAGG)n信号在大多数被检测的物种中几乎检测不到。本文报道了五核苷酸重复序列(TTGGG)n作为蜻蜓和豆娘的典型端粒重复序列。基于10种蜥目动物的基因组信息,(TTGGG)n被认为是最有可能的端粒重复序列的候选者。利用荧光原位杂交(FISH)技术,在12种蜻蜓和豆豆蝇的染色体末端检测到清晰的TTGGG n信号。通过对63种蛇属动物的Southern杂交,大多数蛇属动物均检测到TTGGG - n信号,表明现存蛇属动物共同祖先的端粒重复序列为TTGGG - n。值得注意的是,有少数齿蛙属的(TTGGG)n信号微弱或不存在,这表明端粒重复序列在齿蛙属中反复分化,甚至在属(如Sympetrum)中也是如此。我们还在蜻蜓和豆娘身上发现了端粒酶基因,在一些物种中,可能存在两个以上的端粒酶基因。总的来说,本研究证明了新的端粒重复序列的祖先习得及其在蛇蜥中的重复改变。本文章由计算机程序翻译,如有差异,请以英文原文为准。
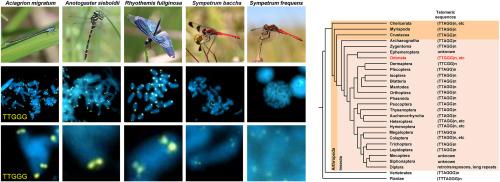
Acquisition and repeated alteration of (TTGGG)n telomeric repeats in Odonata (dragonflies and damselflies)
Chromosome ends of most eukaryotes are composed of simple telomeric repeats. For arthropods, TTAGG pentanucleotide repeats, (TTAGG)n has been considered as the ancestral telomeric repeat. However, in the order Odonata, the earliest diverged group in insects that contains dragonflies and damselflies, (TTAGG)n signal has been almost undetectable in most examined species. Here, we report the pentanucleotide repeat (TTGGG)n as the typical telomeric repeat sequence for Odonata (dragonflies and damselflies). Based on genomic information from ten Odonata species, (TTGGG)n was considered the most likely candidate for telomeric repeat sequences. By fluorescence in situ hybridization (FISH) using 12 Odonata species, clear (TTGGG)n signals were detected at the chromosome ends in both dragonflies and damselflies. By Southern hybridization using 63 Odonata species, strong (TTGGG)n signals were detected from the majority of species covering all three suborders of Odonata, indicating that the telomeric repeat of the common ancestor of extant Odonata is (TTGGG)n. Notably, there were a few Odonata species in which (TTGGG)n signals were faint or absent, suggesting that the telomeric repeat sequence has repeatedly diverged in Odonata, even within genera such as Sympetrum. We also identified telomerase genes in both dragonflies and damselflies, and in some species, more than two telomerase genes are suggested to be present. Overall, this study demonstrates the ancestral acquisition of novel telomeric repeats and their repeated alteration in Odonata.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
7.40
自引率
5.30%
发文量
105
审稿时长
40 days
期刊介绍:
This international journal publishes original contributions and mini-reviews in the fields of insect biochemistry and insect molecular biology. Main areas of interest are neurochemistry, hormone and pheromone biochemistry, enzymes and metabolism, hormone action and gene regulation, gene characterization and structure, pharmacology, immunology and cell and tissue culture. Papers on the biochemistry and molecular biology of other groups of arthropods are published if of general interest to the readership. Technique papers will be considered for publication if they significantly advance the field of insect biochemistry and molecular biology in the opinion of the Editors and Editorial Board.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


