解码土壤生物群系的高分辨率脂质组学:改进的脂质注释、定量和对气候胁迫的响应
IF 10.3
1区 农林科学
Q1 SOIL SCIENCE
引用次数: 0
摘要
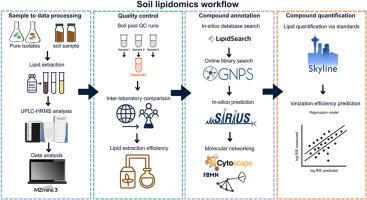
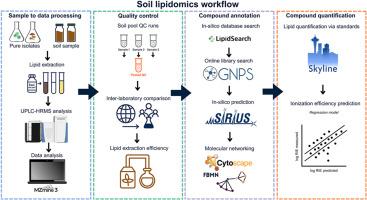
土壤生态系统孕育着多种生物群落,包括古细菌、细菌、真菌、原生生物、植物和土壤动物,它们共同驱动着重要的地下生态系统过程,如养分循环、土壤碳储存和气候调节。目前基于基因的方法提供了最大的分类深度,但是半定量的,并且是探针设计的目标。它们没有覆盖生命之树的整个呼吸,因此还没有完全反映土壤食物网的复杂性。相反,基于脂肪酸的方法提供了微生物群落及其活性的定量见解,但在多细胞生物的分类深度和覆盖范围方面受到限制。我们介绍了一个集成的高分辨率脂组学工作流程,专门设计用于表征土壤生物的广泛多样性。我们的管道结合了从复杂土壤基质中提取脂质,液相色谱-高分辨率Orbitrap质谱,严格的质量控制和多层次的脂质注释策略。此外,我们提出了一个定量的结构-性质关系模型来预测脂质电离效率,为未来在不使用化学标准的情况下改进脂质定量奠定了基础。应用这种方法,我们检测到~ 17000个脂质特征,我们可以注释~ 4800个脂质化合物,与传统方法相比,显著扩大了覆盖范围。脂质谱能有效区分细菌、真菌、植物和藻类等生物,强调了该方法识别生物特异性脂质特征的能力。此外,在模拟气候变化(未来气候和干旱)下的土壤中测试工作流揭示了膜和储存脂质的微妙但有生态意义的变化,突出了脂质体组成对环境胁迫的敏感性。我们的综合脂质组学方法大大推进了脂质注释、定量和生态解释,为生物标志物的发现开辟了新的途径,并提高了对土壤生物群系对环境扰动的响应的理解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
High-resolution lipidomics for decoding the soil biome: Improved lipid annotation, quantitation, and response to climate stress
The soil ecosystem harbors diverse biological communities, including archaea, bacteria, fungi, protists, plants, and soil fauna, that collectively drive essential belowground ecosystem processes such as nutrient cycling, soil carbon storage, and climate regulation. Current gene-based approaches offer greatest taxonomic depth but are semiquantitative and targeted by probe design. They do not cover the whole breath of the tree of life and therefore do not yet fully reflect the complexity of soil food webs. Conversely, fatty acid-based methods provide quantitative insights into microbial communities and their activity but are limited in taxonomic depth and coverage of multicellular organisms. We introduce an integrative high-resolution lipidomics workflow specifically designed to characterize the wide diversity of soil organisms. Our pipeline combines lipid extraction from complex soil matrices with liquid chromatography–high-resolution Orbitrap mass spectrometry, rigorous quality controls, and multitiered lipid annotation strategies. Additionally, we present a quantitative structure–property relationship model to predict lipid ionization efficiencies, providing a foundation for future improvements in lipid quantification without the use of chemical standards. Applying this approach, we detected ∼17,000 lipid features of which we could annotate ∼4800 lipid compounds, significantly expanding the coverage compared with the conventional methods. Lipid profiles effectively distinguish organisms such as bacteria, fungi, plants, and algae, underscoring the ability of this method to identify organism-specific lipid signatures. Furthermore, testing the workflow in soils subjected to simulated climate change (future climate and drought) revealed subtle but ecologically meaningful shifts in membrane and storage lipids, highlighting lipidome compositional sensitivity to environmental stress. Our integrated lipidomics approach substantially advances lipid annotation, quantification, and ecological interpretation, opening new avenues for biomarker discovery and improved understanding of soil biome responses to environmental perturbations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Soil Biology & Biochemistry
农林科学-土壤科学
CiteScore
16.90
自引率
9.30%
发文量
312
审稿时长
49 days
期刊介绍:
Soil Biology & Biochemistry publishes original research articles of international significance focusing on biological processes in soil and their applications to soil and environmental quality. Major topics include the ecology and biochemical processes of soil organisms, their effects on the environment, and interactions with plants. The journal also welcomes state-of-the-art reviews and discussions on contemporary research in soil biology and biochemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


