真菌tRNA连接酶Trl1与RNA的结构揭示了保守的底物结合原则
IF 10.1
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
RNA连接酶在RNA加工和成熟过程中起着至关重要的作用,包括tRNA剪接、RNA修复和未折叠蛋白反应(UPR)。在真菌和植物中,tRNA连接酶Trl1催化tsen切割的前tRNA外显子半部分的连接。在UPR期间,Trl1还作为连接酶参与非常规的HAC1 mRNA剪接。最后的连接步骤由n端腺苷酸转移酶结构域(连接酶;闲逛)。外显子末端在连接反应中的空间排列仍然是难以捉摸的。本文报道了嗜热毛毛菌Trl1-LIG与trna衍生底物配合物的晶体结构。我们的结构代表了激活的RNA中间体的快照,并定义了保守的底物结合界面。潜在的酶-底物相互作用揭示了腺苷基转移酶共享的底物结合原理。此外,我们确定了RNA末端特异性的决定因素以及Trl1-LIG亚结构域在连接酶激活、底物结合和磷酸化转移过程中的特定作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。

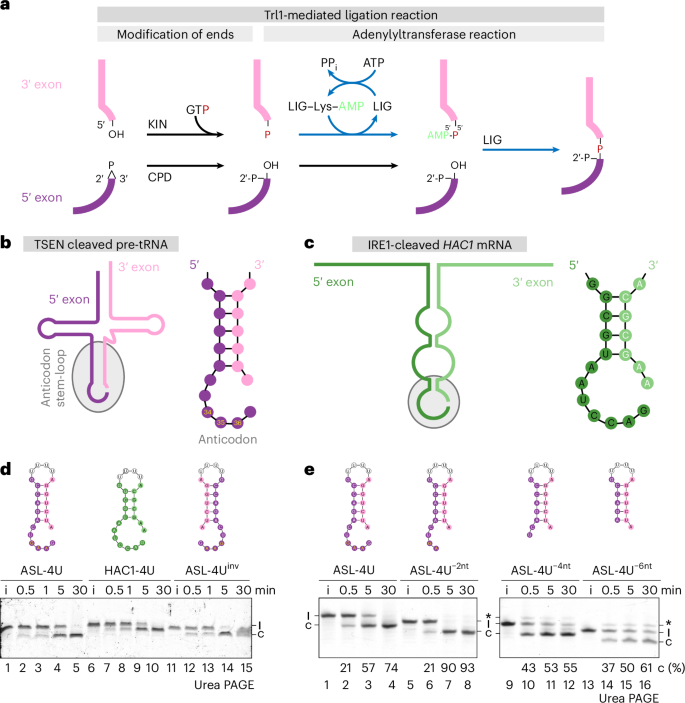
Structure of fungal tRNA ligase Trl1 with RNA reveals conserved substrate-binding principles
RNA ligases play a vital role in RNA processing and maturation, including tRNA splicing, RNA repair and the unfolded protein response (UPR). In fungi and plants, the tripartite tRNA ligase Trl1 catalyzes the joining of TSEN-cleaved pre-tRNA exon halves. Trl1 also functions as ligase in the non-conventional HAC1 mRNA splicing during the UPR. The final ligation step is performed by the N-terminal adenylyltransferase domain (ligase; LIG). The spatial arrangement of the exon ends during the ligation reaction has remained elusive. Here we report the crystal structure of Chaetomium thermophilum Trl1-LIG in complex with a tRNA-derived substrate. Our structure represents a snapshot of the activated RNA intermediate and defines the conserved substrate-binding interface. The underlying enzyme-substrate interplay reveals a substrate-binding principle shared by adenylyltransferases. Moreover, we identify the determinants of RNA end specificity as well as the specific roles of Trl1-LIG’s subdomains during ligase activation, substrate binding and phosphoryl transfer. Köhler et al. present the crystal structure of fungal tRNA ligase Trl1-LIG bound to an activated RNA substrate, providing key insights into conserved substrate binding and activation, enzyme specificity and a tRNA substrate coordination model.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


