纳米针可以实现活组织的时空脂质组学
IF 34.9
1区 材料科学
Q1 MATERIALS SCIENCE, MULTIDISCIPLINARY
引用次数: 0
摘要
空间生物学通过绘制组织的分子组成来提供高含量的诊断信息。然而,传统的空间生物学方法通常需要非生物样本,限制了时间分析。在这里,为了解决这一限制,我们提出了一种使用多孔硅纳米针从活脑组织中反复收集生物分子的工作流程,并通过解吸电喷雾电离质谱成像绘制脂质分布。该方法保留了原始组织的完整性,同时在纳米针底物上复制了其空间分子轮廓,准确反映了脂质分布和组织形态。对23例人类胶质瘤活检的机器学习分析表明,纳米针取样能够精确分类疾病状态。此外,用替莫唑胺治疗的小鼠胶质瘤的时空分析揭示了脂质组成的时间和治疗依赖性变化。我们的方法实现了非破坏性的时空脂质组学,推进了精准医学的分子诊断。本文章由计算机程序翻译,如有差异,请以英文原文为准。
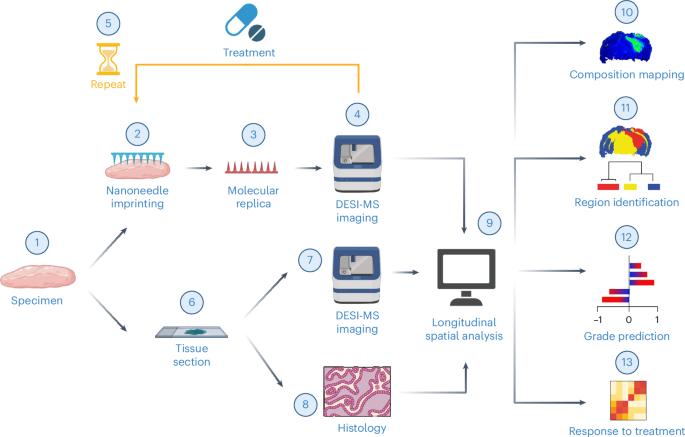

Nanoneedles enable spatiotemporal lipidomics of living tissues
Spatial biology provides high-content diagnostic information by mapping the molecular composition of tissues. However, traditional spatial biology approaches typically require non-living samples, limiting temporal analysis. Here, to address this limitation, we present a workflow using porous silicon nanoneedles to repeatedly collect biomolecules from live brain tissues and map lipid distribution through desorption electrospray ionization mass spectrometry imaging. This method preserves the integrity of the original tissue while replicating its spatial molecular profile on the nanoneedle substrate, accurately reflecting lipid distribution and tissue morphology. Machine learning analysis of 23 human glioma biopsies demonstrated that nanoneedle sampling enables the precise classification of disease states. Furthermore, a spatiotemporal analysis of mouse gliomas treated with temozolomide revealed time- and treatment-dependent variations in lipid composition. Our approach enables non-destructive spatiotemporal lipidomics, advancing molecular diagnostics for precision medicine. Arrays of silicon nanoneedles are used to generate molecular replicas of live brain tissue for longitudinal spatial lipidomic classification via desorption electrospray ionization mass spectrometry imaging of gliomas and to monitor the responses of the tumours to chemotherapy.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature nanotechnology
工程技术-材料科学:综合
CiteScore
59.70
自引率
0.80%
发文量
196
审稿时长
4-8 weeks
期刊介绍:
Nature Nanotechnology is a prestigious journal that publishes high-quality papers in various areas of nanoscience and nanotechnology. The journal focuses on the design, characterization, and production of structures, devices, and systems that manipulate and control materials at atomic, molecular, and macromolecular scales. It encompasses both bottom-up and top-down approaches, as well as their combinations.
Furthermore, Nature Nanotechnology fosters the exchange of ideas among researchers from diverse disciplines such as chemistry, physics, material science, biomedical research, engineering, and more. It promotes collaboration at the forefront of this multidisciplinary field. The journal covers a wide range of topics, from fundamental research in physics, chemistry, and biology, including computational work and simulations, to the development of innovative devices and technologies for various industrial sectors such as information technology, medicine, manufacturing, high-performance materials, energy, and environmental technologies. It includes coverage of organic, inorganic, and hybrid materials.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


