犬猫感染鲍曼不动杆菌和皮蒂不动杆菌多个克隆中abar4型基因组岛和多药耐药质粒的定位
IF 2.6
4区 医学
Q3 INFECTIOUS DISEASES
引用次数: 0
摘要
对泰国(2016-2019)临床产碳青霉烯酶的39株鲍曼不动杆菌(CP-Ab)和2株皮蒂不动杆菌(CP-Ap)基因组进行了系统发育关系和抗微生物药物耐药性基因(ARG)定位分析。茎Resistome和核心基因组序列输入(cgMLST)分析确定不同的参数模式在CP-Ab序列类型(ST) 2 (n = 4)和ST1575 (n = 1)克隆复杂(CC) 2、ST25 (n = 6),ST1576 (n = 1),和ST1581 CC25 (n = 2),ST16 (n = 13),ST23 (n = 2),ST149 (n = 9),ST1093 (n = 1),连同CP-Ap (n = 2)。对每个克隆的1株CP-Ap和15株具有代表性的CP-Ab进行全基因组分析,发现菌株具有1 - 3个与Tn2006和/或abar型基因组岛(GIs)相关的blaOXA-23拷贝。除了CC2的CP-Ab携带AbaR4b外,其他所有STs的染色体上都有AbaR4。在CP-Ab ST1093和CP-Ap菌株的共轭OXA-23质粒上发现了AbaR4。CP-Ab ST149和CC25的共轭OXA-23质粒和多药耐药(MDR)大质粒与CP-Ab ST23的隐质粒pCUVET16-467.1具有共同的祖先。MDR巨型质粒包含一个噬菌体排斥位点(BREX)和tn6172衍生的元件。在ST149的GR38 MDR巨型质粒中发现了含有1类整合子GR38 repM和多个ARGs的Tn6172ISCR2::(ΔISCR2-ΔTn10-MARR)结构,在CC25质粒中形成了AbGRI1结构。这些发现强调了AbaR4在碳青霉烯耐药和不动杆菌质粒进化中的作用,促进了blaOXA-23在来自伴侣动物的临床CP-Ab和CP-Ap菌株的多个克隆中的传播和广泛耐药的发展。本文章由计算机程序翻译,如有差异,请以英文原文为准。
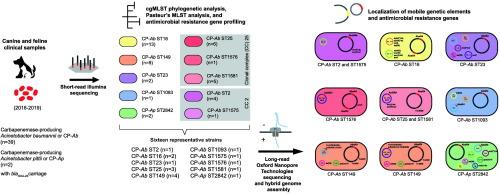
Localization of AbaR4-type genomic islands and multidrug resistance plasmids in multiple Acinetobacter baumannii clones and Acinetobacter pittii from infections of dogs and cats
The genomes of 39 clinical carbapenemase-producing A. baumannii (CP-Ab) and 2 A. pittii (CP-Ap) isolates from dogs and cats in Thailand (2016–2019) were analyzed for phylogenetic relationship and antimicrobial resistance gene (ARG) localization. Resistome and core genome multilocus sequence typing (cgMLST) analysis identified distinct ARG patterns within CP-Ab sequence type (ST) 2 (n = 4) and ST1575 (n = 1) of clonal complex (CC) 2, ST25 (n = 6), ST1576 (n = 1), and ST1581 (n = 2) of CC25, ST16 (n = 13), ST23 (n = 2), ST149 (n = 9), ST1093 (n = 1), along with CP-Ap (n = 2). Complete genome analysis of one CP-Ap strain and 15 representative CP-Ab strains from each clone identified strains with one to three copies of blaOXA-23 associated with Tn2006 and/or AbaR-type genomic islands (GIs). AbaR4 was chromosomally located in all other STs except CP-Ab of CC2, which carried AbaR4b. AbaR4 was found on conjugative blaOXA-23-containing plasmids in CP-Ab ST1093 and CP-Ap strains. Conjugative blaOXA-23-containing plasmids and multidrug resistance (MDR) mega-plasmids in CP-Ab ST149 and CC25 shared ancestry with the cryptic plasmid pCUVET16–467.1 in CP-Ab ST23. MDR mega-plasmids contained a bacteriophage exclusion (BREX) locus and Tn6172-derived elements. A Tn6172ISCR2::(ΔISCR2-ΔTn10-MARR) structure containing GR38 repM, a class 1 integron, and multiple ARGs were found in R3_T60 (GR38) MDR mega-plasmids of ST149 and formed an AbGRI1 structure in CC25 plasmids. These findings highlight the role of AbaR4 in carbapenem resistance and the evolution of Acinetobacter plasmids, facilitating the blaOXA-23 dissemination and the development of extensive drug resistance in multiple clones of clinical CP-Ab and CP-Ap strains originating from companion animals.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Infection Genetics and Evolution
医学-传染病学
CiteScore
8.40
自引率
0.00%
发文量
215
审稿时长
82 days
期刊介绍:
(aka Journal of Molecular Epidemiology and Evolutionary Genetics of Infectious Diseases -- MEEGID)
Infectious diseases constitute one of the main challenges to medical science in the coming century. The impressive development of molecular megatechnologies and of bioinformatics have greatly increased our knowledge of the evolution, transmission and pathogenicity of infectious diseases. Research has shown that host susceptibility to many infectious diseases has a genetic basis. Furthermore, much is now known on the molecular epidemiology, evolution and virulence of pathogenic agents, as well as their resistance to drugs, vaccines, and antibiotics. Equally, research on the genetics of disease vectors has greatly improved our understanding of their systematics, has increased our capacity to identify target populations for control or intervention, and has provided detailed information on the mechanisms of insecticide resistance.
However, the genetics and evolutionary biology of hosts, pathogens and vectors have tended to develop as three separate fields of research. This artificial compartmentalisation is of concern due to our growing appreciation of the strong co-evolutionary interactions among hosts, pathogens and vectors.
Infection, Genetics and Evolution and its companion congress [MEEGID](http://www.meegidconference.com/) (for Molecular Epidemiology and Evolutionary Genetics of Infectious Diseases) are the main forum acting for the cross-fertilization between evolutionary science and biomedical research on infectious diseases.
Infection, Genetics and Evolution is the only journal that welcomes articles dealing with the genetics and evolutionary biology of hosts, pathogens and vectors, and coevolution processes among them in relation to infection and disease manifestation. All infectious models enter the scope of the journal, including pathogens of humans, animals and plants, either parasites, fungi, bacteria, viruses or prions. The journal welcomes articles dealing with genetics, population genetics, genomics, postgenomics, gene expression, evolutionary biology, population dynamics, mathematical modeling and bioinformatics. We also provide many author benefits, such as free PDFs, a liberal copyright policy, special discounts on Elsevier publications and much more. Please click here for more information on our author services .
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


