不同盐水平下抗生素耐药基因的偶联效应:单因素和模拟研究的见解
IF 6.9
2区 环境科学与生态学
Q1 ENVIRONMENTAL SCIENCES
引用次数: 0
摘要
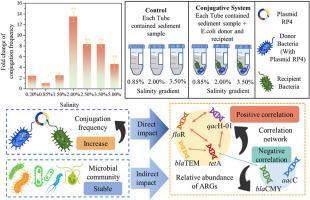
抗生素耐药基因(ARGs)在盐度波动下的传播机制仍然知之甚少,尽管它们对环境抗性生态学具有重要意义。本研究通过受控的单因素实验和模拟沉积物微观环境,系统地解耦了盐度驱动的共轭动力学。对照偶联实验显示了阈值依赖性的响应,在盐度为2.00 %时,RP4质粒转移频率达到峰值(比0.85%的对照增加4.58 - 13.51倍,p <;0.01),与活性氧(ROS)介导的SOS通路激活机制相关。在模拟沉积物系统中,盐度梯度驱动宿主特异性ARGs富集,在微咸条件下(盐度为2.00%),质粒携带的tetA和blaTEM丰度增加了1.49-4.39倍。多药耐药基因floR、qacH-01呈现协同扩散模式(r = 0.77 ~ 0.94, p <;盐度为3.50%时,弯曲菌门和螺旋藻门成为ARGs的优势储层(富集程度为对照的2.05-3.17倍)。虽然外源抗生素耐药菌(ARB)的引入略微降低了α-多样性,但门水平的群落结构保持稳定。盐度优先抑制稀有类群,通过生态位重构放大ARGs共现网络。这些发现表明盐度是ARGs传播的双重调节因子,通过氧化应激途径直接增强结合,并通过微生物宿主选择间接重塑抗性景观。结果强调了将盐度梯度纳入ARGs风险评估的必要性,特别是在潮汐波动可能加剧抗性传播的沿海生态系统中。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Conjugation effects on antibiotic resistance genes at various salt Levels: Insights from single-factor and simulation study
The dissemination mechanisms of antibiotic resistance genes (ARGs) under salinity fluctuations remain poorly understood, despite their critical implications for environmental resistance ecology. This study systematically decoupled salinity-driven conjugation dynamics through controlled single-factor experiments and simulated sediment microcosms. Controlled conjugation assays revealed a threshold-dependent response, with RP4 plasmid transfer frequencies peaking at 2.00 % salinity (4.58–13.51-fold increase vs. 0.85 % control, p < 0.01), mechanistically linked to reactive oxygen species (ROS)-mediated SOS pathway activation. In simulated sediment systems, salinity gradients drove host-specific ARGs enrichment, with plasmid-borne tetA and blaTEM abundances increasing 1.49–4.39 fold under brackish conditions (2.00 % salinity). Multidrug resistance genes floR, qacH-01 exhibited synergistic diffusion patterns (r = 0.77–0.94, p < 0.05), while salt-tolerant phyla Campylobacterota and Spirochaetota became dominant ARGs reservoirs at 3.50 % salinity (2.05–3.17 fold enrichment vs. controls). Although exogenous antibiotic resistance bacteria (ARB) introduction marginally reduced α-diversity, phylum-level community structure remained stable. Salinity preferentially suppressed rare taxa, amplifying ARGs co-occurrence networks through niche restructuring. These findings establish salinity as a dual regulator of ARGs dissemination, directly enhancing conjugation via oxidative stress pathways and indirectly reshaping resistance landscapes through microbial host selection. The results underscore the necessity of integrating salinity gradients into ARGs risk assessments, particularly in coastal ecosystems where tidal fluctuations may potentiate resistance propagation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Emerging Contaminants
Medicine-Public Health, Environmental and Occupational Health
CiteScore
10.00
自引率
6.70%
发文量
35
审稿时长
44 days
期刊介绍:
Emerging Contaminants is an outlet for world-leading research addressing problems associated with environmental contamination caused by emerging contaminants and their solutions. Emerging contaminants are defined as chemicals that are not currently (or have been only recently) regulated and about which there exist concerns regarding their impact on human or ecological health. Examples of emerging contaminants include disinfection by-products, pharmaceutical and personal care products, persistent organic chemicals, and mercury etc. as well as their degradation products. We encourage papers addressing science that facilitates greater understanding of the nature, extent, and impacts of the presence of emerging contaminants in the environment; technology that exploits original principles to reduce and control their environmental presence; as well as the development, implementation and efficacy of national and international policies to protect human health and the environment from emerging contaminants.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


