迈向一个完整的噬菌体尾部纤维结构图谱
IF 12.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
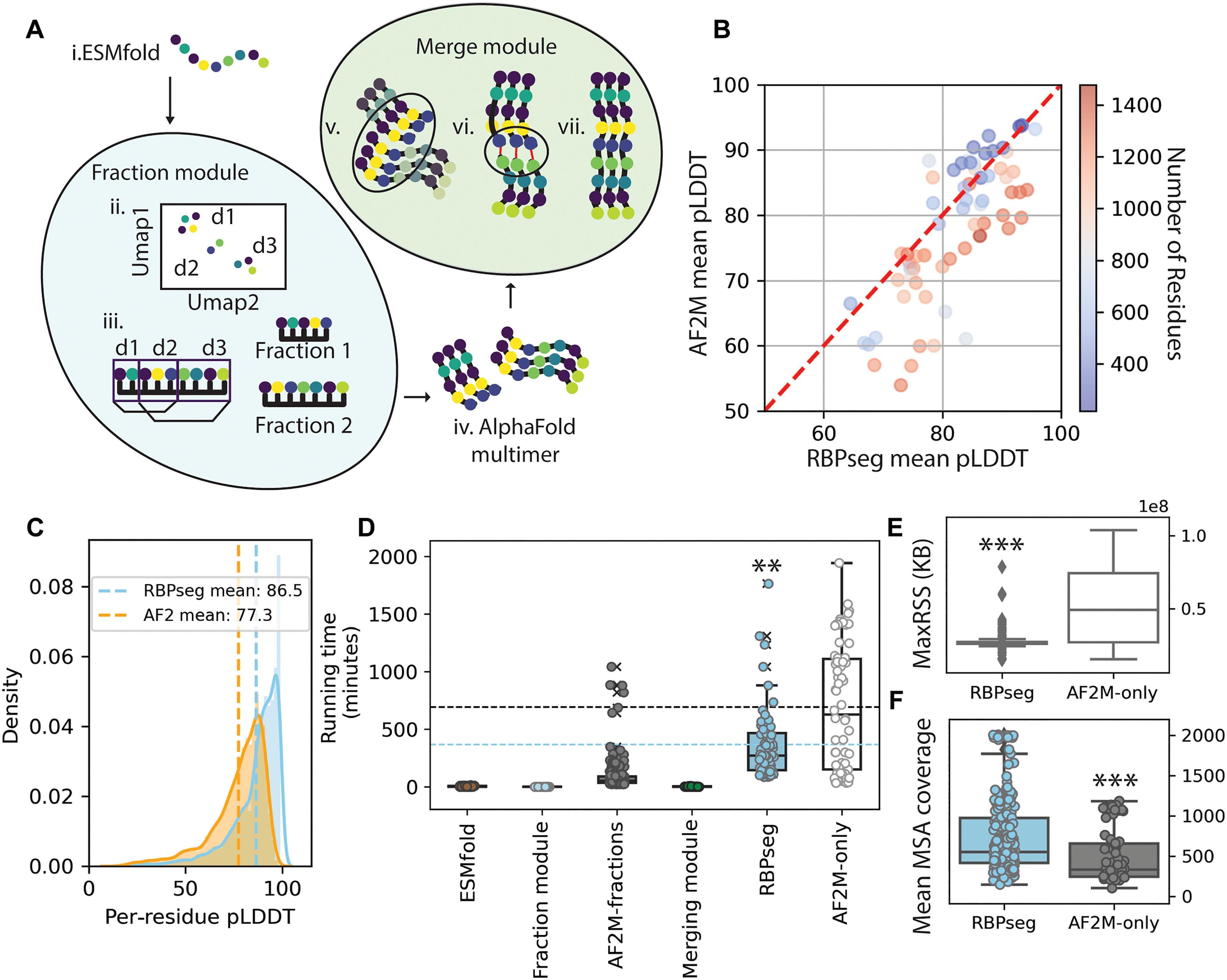
噬菌体利用受体结合蛋白(rbp)附着在细菌宿主上,但人们对其序列和结构多样性知之甚少。尾纤维是一类主要的rbp,是一种细长的、柔性的三聚体蛋白,使其全长结构难以在实验中解析。基于深度学习的蛋白质结构预测的进展,如alphafold2 - multitimer (AF2M)和ESMFold,为研究这些具有挑战性的蛋白质提供了机会。在这里,我们引入了RBPseg,一种将单体ESMFold预测与基于结构的域识别方法相结合的方法,将尾部纤维序列划分为可管理的部分,以便使用AF2M进行高置信度建模。利用这种方法,我们生成了完整的尾巴纤维模型,并通过单粒子低温电子显微镜验证了来自三个噬菌体的五根纤维。67根纤维的结构分类确定了16个不同的类别和89个域,揭示了模块化、收敛、发散和域交换的模式。我们的研究结果表明,这些结构类别至少占已知尾纤维宇宙的24%,为它们的进化和功能提供了关键的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
RBPseg: Toward a complete phage tail fiber structure atlas
Bacteriophages use receptor-binding proteins (RBPs) to adhere to bacterial hosts, yet their sequence and structural diversity remain poorly understood. Tail fibers, a major class of RBPs, are elongated and flexible trimeric proteins, making their full-length structures difficult to resolve experimentally. Advances in deep learning–based protein structure prediction, such as AlphaFold2-multimer (AF2M) and ESMFold, provide opportunities for studying these challenging proteins. Here, we introduce RBPseg, a method that combines monomeric ESMFold predictions with a structural-based domain identification approach, to divide tail fiber sequences into manageable fractions for high-confidence modeling with AF2M. Using this approach, we generated complete tail fiber models, validated by single-particle cryo–electron microscopy of five fibers from three phages. A structural classification of 67 fibers identified 16 distinct classes and 89 domains, revealing patterns of modularity, convergence, divergence, and domain swapping. Our findings suggest that these structural classes represent at least 24% of the known tail fiber universe, providing key insights into their evolution and functionality.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Science Advances
综合性期刊-综合性期刊
CiteScore
21.40
自引率
1.50%
发文量
1937
审稿时长
29 weeks
期刊介绍:
Science Advances, an open-access journal by AAAS, publishes impactful research in diverse scientific areas. It aims for fair, fast, and expert peer review, providing freely accessible research to readers. Led by distinguished scientists, the journal supports AAAS's mission by extending Science magazine's capacity to identify and promote significant advances. Evolving digital publishing technologies play a crucial role in advancing AAAS's global mission for science communication and benefitting humankind.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


