淀粉样蛋白形成片段对人类CPEB3朊病毒功能调控的结构见解
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
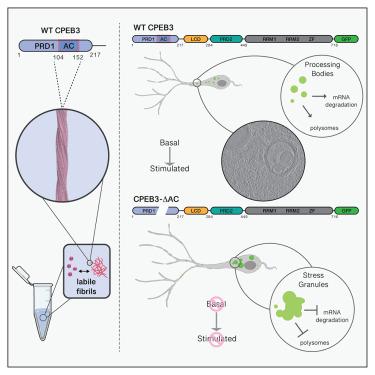
胞质聚腺苷化元件结合蛋白-3 (CPEB3)是一种功能性朊病毒,被认为可以调节蛋白质合成并促进神经元长期记忆的巩固。我们报道了从人CPEB3的第一个朊病毒样结构域(hCPEB3)体外培养的淀粉样原纤维的冷冻电镜(cro - em)结构,揭示了它们有序的49个残基核心,从L103到F151。与野生型CPEB3相比,缺乏该片段的CPEB3在细胞中凝聚成异常点,定位于远离休眠p体和应激颗粒的位置,并且缺乏影响神经元中蛋白质合成的能力。荧光引导冷冻聚焦离子束(cro - fib)铣削和冷冻电子断层扫描(cro - et)应用于表达CPEB3的神经元细胞,发现CPEB3- gfp信号来自多泡体(MVBs)、海绵状多层膜室和束状细丝中富集的片层,表明细胞处于诱导应激状态。因此,表达野生型CPEB3的细胞比没有淀粉样蛋白核心的表达CPEB3的细胞存活率低,这表明人类CPEB3可能需要调节来克服其在细胞中自组装相关的缺陷。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Structural insights into functional regulation of the human CPEB3 prion by an amyloid-forming segment
The cytoplasmic polyadenylation-element-binding-protein-3 (CPEB3) is a functional prion thought to modulate protein synthesis and enable consolidation of long-term memory in neurons. We report a cryoelectron microscopy (cryo-EM) structure of amyloid fibrils grown in vitro from the first prion-like domain of human CPEB3 (hCPEB3), revealing their ordered 49-residue core, spanning L103 to F151. CPEB3 lacking that segment coalesces into abnormal puncta in cells compared to wild-type CPEB3, localizes away from dormant p-bodies and toward stress granules, and lacks the ability to influence protein synthesis in neurons. Fluorescence-guided cryo-focused ion beam (cryo-FIB) milling and cryo-electron tomography (cryo-ET) applied to neuronal cells expressing CPEB3 reveal CPEB3-GFP signal from lamellae enriched in multivesicular bodies (MVBs), cavernous multilamellar compartments, and bundled filaments, suggesting a state of induced cellular stress. Accordingly, cells expressing wild-type CPEB3 are less viable than those expressing CPEB3 without its amyloid core, suggesting human CPEB3 regulation may be required to overcome the liability associated with its self-assembly in cells.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


