利用深度学习预测超声心动图视频中的心血管磁共振结果。
IF 6
2区 医学
Q1 CARDIAC & CARDIOVASCULAR SYSTEMS
Journal of the American Society of Echocardiography
Pub Date : 2025-09-01
DOI:10.1016/j.echo.2025.05.016
引用次数: 0
摘要
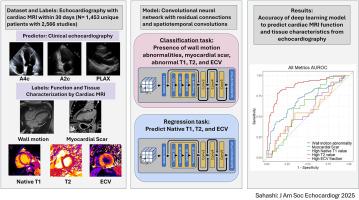
背景:超声心动图是评估心脏结构和功能最常用的方法。虽然心脏磁共振(CMR)成像不太容易获得,但CMR可以提供独特的组织特征,包括晚期钆增强(LGE)、T1和T2定位以及与组织纤维化、浸润和炎症相关的细胞外体积(ECV)。深度学习已被证明可以揭示临床医生未认识到的发现,然而,目前尚不清楚基于cmr的组织特征是否可以使用深度学习从超声心动图视频中获得。目的:评估应用于超声心动图的深度学习模型检测cmr特异性参数的性能,包括LGE的存在,以及异常的T1、T2或ECV。方法:在一项回顾性单中心研究中,纳入了30天内进行cmr和超声心动图检查的成年患者。基于视频的卷积神经网络在超声心动图视频上进行训练,以预测cmr衍生的标签,包括LGE的存在,以及超声心动图视图上异常的T1、T2或ECV。该模型还被训练来预测壁面运动异常(WMA)的存在/不存在,作为模型功能的正控制。模型的性能在一个不用于训练的测试数据集中进行评估。结果:研究人群包括1453名成年患者(平均年龄56±18岁,42%为女性),在CMR后2天(四分位数间隔为2天至6天)进行了2556次配对超声心动图研究。该模型对阳性对照WMA的存在有较高的预测能力(AUC 0.873 [95%CI 0.816-0.922])。然而,该模型无法可靠地检测LGE (AUC 0.699[0.613-0.780])、异常原生T1 (AUC 0.614[0.500-0.715])、T2 0.553[0.20 -0.692]或ECV 0.564[0.455-0.691])的存在。结论:应用于超声心动图的深度学习准确地识别了基于CMR的WMA,但无法预测组织特征,这表明超声视频中可能不存在这些组织特征的信号,并且在心脏病学中使用CMR进行组织表征仍然是必不可少的。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Using Deep Learning to Predict Cardiovascular Magnetic Resonance Findings From Echocardiographic Videos
Background
Echocardiography is the most common modality for assessing cardiac structure and function. Although cardiac magnetic resonance (CMR) imaging is less accessible, it can provide unique tissue characterization, including late gadolinium enhancement (LGE), T1 and T2 mapping, and extracellular volume (ECV), which are associated with tissue fibrosis, infiltration, and inflammation. Deep learning has been shown to uncover findings not recognized by clinicians, but it is unknown whether CMR-based tissue characteristics can be derived from echocardiographic videos using deep learning. The aim of this study was to assess the performance of a deep learning model applied to echocardiography to detect CMR-specific parameters, including LGE presence and abnormal T1, T2, or ECV.
Methods
In a retrospective single-center study, adult patients with CMR and echocardiographic studies within 30 days were included. A video-based convolutional neural network was trained on echocardiographic videos to predict CMR-derived labels, including LGE presence and abnormal T1, T2, or ECV across echocardiographic views. The model was also trained to predict the presence or absence of wall motion abnormality (WMA) as a positive control for model function. The model performance was evaluated in a held-out test data set not used for training.
Results
The study population included 1,453 adult patients (mean age, 56 ± 18 years; 42% women) with 2,556 paired echocardiographic studies occurring at a median of 2 days after CMR (interquartile range, 2 days before to 6 days after). The model had high predictive capability for the presence of WMA (area under the curve [AUC] = 0.873; 95% CI, 0.816-0.922), which was used for positive control. However, the model was unable to reliably detect the presence of LGE (AUC = 0.699; 95% CI, 0.613-0.780) and abnormal native T1 (AUC = 0.614; 95% CI, 0.500-0.715), T2 (AUC = 0.553; 95% CI, 0.420-0.692), or ECV (AUC = 0.564; 95% CI, 0.455-0.691).
Conclusions
Deep learning applied to echocardiography accurately identified CMR-based WMA but was unable to predict tissue characteristics, suggesting that signal for these tissue characteristics may not be present within ultrasound videos and that the use of CMR for tissue characterization remains essential within cardiology.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
9.50
自引率
12.30%
发文量
257
审稿时长
66 days
期刊介绍:
The Journal of the American Society of Echocardiography(JASE) brings physicians and sonographers peer-reviewed original investigations and state-of-the-art review articles that cover conventional clinical applications of cardiovascular ultrasound, as well as newer techniques with emerging clinical applications. These include three-dimensional echocardiography, strain and strain rate methods for evaluating cardiac mechanics and interventional applications.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


