基于selex的适体生产:霍乱检测的关键步骤
IF 0.9
Q4 GENETICS & HEREDITY
引用次数: 0
摘要
霍乱弧菌(V. cholerae)是造成全世界腹泻死亡人数第二高的疾病。这种微生物与霍乱暴发密切相关,无论是大流行还是流行。因此,早期快速检测方法对于预防疾病的传播至关重要。霍乱毒素(Ctx)是霍乱弧菌的一种毒力因子,在该细菌的发病机制中起着主要和必要的作用。适配体是一种单链(ss)折叠RNA或ssDNA,可以结合并检测各种核酸和非核酸分子,具有高亲和力和特异性,在生物标志物发现,诊断,成像和靶向治疗等各种应用中提供了抗体的有希望的替代品。这些适体是通过一个称为配体系统进化的过程通过指数富集(SELEX)选择的。本研究的目的是通过不对称PCR制备适配体,不对称PCR的最佳条件为引物比例为1:80,循环20次,温度为57℃。运行11轮SELEX后,分离合适的适配体。计算最佳CtxB适配体的检测限(LoD)和解离常数(KD),以评估其特异性和亲和力。测定LoD为100±20 pg/ml, KD为320±60 pM。本文章由计算机程序翻译,如有差异,请以英文原文为准。
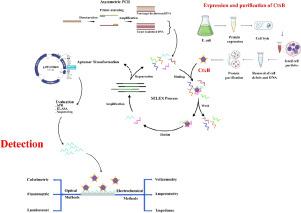
SELEX-based aptamer production: A key step in cholera detection
Vibrio cholerae (V. cholerae) is responsible for the second-highest number of deaths worldwide due to diarrhea. This microorganism is closely associated with cholera outbreaks, both pandemic and epidemic. Therefore, an early rapid detection assay is crucial to prevent the disease's spread. Cholera toxin (Ctx) is a V. cholerae virulence factor that plays a primary and essential role in the pathogenesis of this bacterium. Aptamers, a single-stranded (ss) folded RNA or ssDNA that can bind and detect various nucleic and non-nucleic acid molecules with high affinity and specificity, offer a promising alternative to antibodies in various applications such as biomarker discovery, diagnosis, imaging, and targeted therapy. These aptamers are selected through a process called systematic evolution of ligands by exponential enrichment (SELEX). The present study aimed to produce aptamers by asymmetric PCR, the best conditions obtained for asymmetric PCR included the ratio of 1:80 primers, 20 cycles, and a temperature of 57 °C. After running 11 rounds of SELEX, appropriate aptamers were isolated. The limit of detection (LoD) and dissociation constant (KD) of the best aptamer for CtxB were calculated for specificity and affinity. The LoD was measured as 100 ± 20 pg/ml, and the KD was calculated as 320 ± 60 pM.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Gene Reports
Biochemistry, Genetics and Molecular Biology-Genetics
CiteScore
3.30
自引率
7.70%
发文量
246
审稿时长
49 days
期刊介绍:
Gene Reports publishes papers that focus on the regulation, expression, function and evolution of genes in all biological contexts, including all prokaryotic and eukaryotic organisms, as well as viruses. Gene Reports strives to be a very diverse journal and topics in all fields will be considered for publication. Although not limited to the following, some general topics include: DNA Organization, Replication & Evolution -Focus on genomic DNA (chromosomal organization, comparative genomics, DNA replication, DNA repair, mobile DNA, mitochondrial DNA, chloroplast DNA). Expression & Function - Focus on functional RNAs (microRNAs, tRNAs, rRNAs, mRNA splicing, alternative polyadenylation) Regulation - Focus on processes that mediate gene-read out (epigenetics, chromatin, histone code, transcription, translation, protein degradation). Cell Signaling - Focus on mechanisms that control information flow into the nucleus to control gene expression (kinase and phosphatase pathways controlled by extra-cellular ligands, Wnt, Notch, TGFbeta/BMPs, FGFs, IGFs etc.) Profiling of gene expression and genetic variation - Focus on high throughput approaches (e.g., DeepSeq, ChIP-Seq, Affymetrix microarrays, proteomics) that define gene regulatory circuitry, molecular pathways and protein/protein networks. Genetics - Focus on development in model organisms (e.g., mouse, frog, fruit fly, worm), human genetic variation, population genetics, as well as agricultural and veterinary genetics. Molecular Pathology & Regenerative Medicine - Focus on the deregulation of molecular processes in human diseases and mechanisms supporting regeneration of tissues through pluripotent or multipotent stem cells.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


