罗伊氏乳酸杆菌精氨酸脱亚胺酶(LrADI)的克隆及酶学特性研究
IF 0.9
Q4 GENETICS & HEREDITY
引用次数: 0
摘要
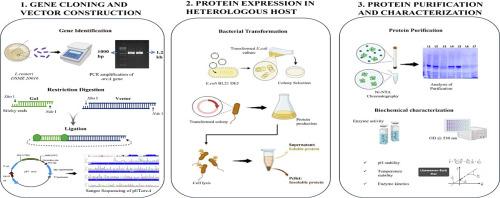
精氨酸脱亚胺酶(Arginine de亚胺酶,ADI)是一种通过胍脱胺将l -精氨酸水解成l -瓜氨酸和氨的弧线降解酶。它属于胍基修饰酶(GME)超家族,在微生物ADI途径中起着至关重要的作用。本研究从罗伊氏乳酸杆菌DSM 20016中克隆并表达了adi编码基因arcA。开放阅读框(ORF)包含1233个碱基对,编码411个氨基酸的蛋白质。构建重组pET28b-arcA表达载体,转化大肠杆菌BL21 (DE3)进行外源表达。用IPTG诱导蛋白表达,用Ni-NTA组氨酸标记亲和层析纯化重组ADI (LrADI)。生化表征表明,最佳反应温度为40 °C, pH为6。动力学分析表明,该反应的Michaelis常数为1.63 mM,最大反应速度(Vmax)为1.08 mM/min。SDS-PAGE证实分子质量为~46 kDa。这些发现为LrADI在制药行业的酶学特性和潜在应用提供了见解,特别是在癌症治疗中,因为它能够消耗精氨酸-营养不良肿瘤中的精氨酸。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Molecular cloning and enzymatic characterization of arginine deiminase (LrADI) from Limosilactobacillus reuteri DSM 20016: A candidate for cancer metabolic therapy
Arginine deiminase (ADI) is an arc-degrading enzyme that hydrolyzes L-arginine into L-citrulline and ammonia through guanidine deamination. It belongs to the guanidino group modifying enzymes (GME) superfamily, which plays a crucial role in the microbial ADI pathway. In this study, the ADI-encoding gene arcA from Limosilactobacillus reuteri DSM 20016 was cloned and expressed. The open reading frame (ORF) comprises 1233 base pairs, encodes a protein of 411 amino acids. A recombinant pET28b-arcA expression vector was constructed and transformed into Escherichia coli BL21 (DE3) for heterologous expression. Protein expression was induced with IPTG, and the recombinant ADI (LrADI) was purified using Ni-NTA histidine-tagged affinity chromatography. Biochemical characterization revealed an optimal reaction temperature of 40 °C and a pH optimum of 6. Kinetic analysis showed a Michaelis constant of 1.63 mM and a maximum reaction velocity (Vmax) of 1.08 mM/min. SDS-PAGE confirmed a molecular mass of ~46 kDa. These findings provide insights into the enzymatic properties and potential applications of LrADI in the pharmaceutical industry, particularly in cancer treatment, due to its ability to deplete arginine in arginine-auxotrophic tumors.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Gene Reports
Biochemistry, Genetics and Molecular Biology-Genetics
CiteScore
3.30
自引率
7.70%
发文量
246
审稿时长
49 days
期刊介绍:
Gene Reports publishes papers that focus on the regulation, expression, function and evolution of genes in all biological contexts, including all prokaryotic and eukaryotic organisms, as well as viruses. Gene Reports strives to be a very diverse journal and topics in all fields will be considered for publication. Although not limited to the following, some general topics include: DNA Organization, Replication & Evolution -Focus on genomic DNA (chromosomal organization, comparative genomics, DNA replication, DNA repair, mobile DNA, mitochondrial DNA, chloroplast DNA). Expression & Function - Focus on functional RNAs (microRNAs, tRNAs, rRNAs, mRNA splicing, alternative polyadenylation) Regulation - Focus on processes that mediate gene-read out (epigenetics, chromatin, histone code, transcription, translation, protein degradation). Cell Signaling - Focus on mechanisms that control information flow into the nucleus to control gene expression (kinase and phosphatase pathways controlled by extra-cellular ligands, Wnt, Notch, TGFbeta/BMPs, FGFs, IGFs etc.) Profiling of gene expression and genetic variation - Focus on high throughput approaches (e.g., DeepSeq, ChIP-Seq, Affymetrix microarrays, proteomics) that define gene regulatory circuitry, molecular pathways and protein/protein networks. Genetics - Focus on development in model organisms (e.g., mouse, frog, fruit fly, worm), human genetic variation, population genetics, as well as agricultural and veterinary genetics. Molecular Pathology & Regenerative Medicine - Focus on the deregulation of molecular processes in human diseases and mechanisms supporting regeneration of tissues through pluripotent or multipotent stem cells.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


