改进遗传性听力损失患者STRC基因15q15.3区域复杂重排的检测。
IF 2.5
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
摘要
听力损失(HL)是世界范围内最常见的感觉障碍,GJB2-GJB6连接蛋白改变(DFNB1)是非综合征性听力损失(NSHL)的最常见原因。最近的研究也强调了STRC基因是NSHL的重要贡献者,其发病率可能接近连接蛋白改变。尽管新一代测序技术(NGS)取得了进步,但对于许多NSHL患者来说,分子诊断仍然具有挑战性,这通常是由于分析STRC基因的复杂性。这在很大程度上归因于它位于串联复制区域和同源假基因(STRCP1)的存在,这使得其准确鉴定变得复杂。DFNB16最常见的原因是15q15.3的纯合大连续基因缺失,但其他拷贝数变异(cnv),包括丢失和获得,尚未被很好地描述。通过这些技术的结合,我们提供了来自59个家庭的72名DFNB16患者的STRC变异和诊断的新数据。虽然CKMT1B-STRC-CATSPER2缺失是最常见的改变,但改进的水滴数字PCR (ddPCR)用于改进基因前16个外显子的CNV分析(99.8%与假基因同源)使我们能够识别并更好地定义更高发生率的以前未明确的复杂重排。此外,我们已经确定了c.4837 G > T;p.(Glu1613*)致病变异与CATSPER2-CKMT1A-STRCP1重复之间的直接顺式关联。这些发现强调了ddPCR在识别传统NGS难以检测到的CNVs方面的重要作用,显著提高了诊断水平,并为受影响家庭提供了精确的遗传咨询。本文章由计算机程序翻译,如有差异,请以英文原文为准。
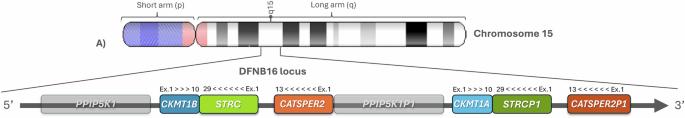
Refining the detection of complex rearrangements in 15q15.3 region involving the STRC gene in hereditary hearing loss patients
Hearing loss (HL) is the most common sensory disability worldwide, with GJB2-GJB6 connexin alterations (DFNB1) being the most frequent causes of non-syndromic hearing loss (NSHL). Recent studies have also highlighted the STRC gene as a significant contributor to NSHL, with its incidence potentially approaching that of connexin alterations. Despite advances in next-generation sequencing (NGS), molecular diagnosis remains challenging for many NSHL patients, often due to the complexity of analyzing the STRC gene. This is largely attributed to its location in a tandemly duplicated region and the presence of a homologous pseudogene (STRCP1), which complicates its accurate identification. The most common cause of DFNB16 is a homozygous large contiguous gene deletion at 15q15.3, but other copy number variants (CNVs), including both losses and gains, have been less well characterized. Through a combination of techniques we present new data on STRC variants and the diagnosis of 72 DFNB16 patients from 59 families. While the CKMT1B-STRC-CATSPER2 deletion is the most frequent alteration, the improvement of droplet-digital PCR (ddPCR) for refining CNV analysis in the first 16 exons of the gene (99,8% homologous with the pseudogene) has allowed us to identify and better define a higher incidence of previously unclarified complex rearrangements. Additionally, we have identified a direct cis association between the c.4837 G > T;p.(Glu1613*) pathogenic variant and the CATSPER2-CKMT1A-STRCP1 duplication. These findings underscore the important role of ddPCR in identifying CNVs that are difficult to detect through conventional NGS, significantly improving diagnosis and enabling precise genetic counseling for affected families.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Human Genetics
生物-遗传学
CiteScore
7.20
自引率
0.00%
发文量
101
审稿时长
4-8 weeks
期刊介绍:
The Journal of Human Genetics is an international journal publishing articles on human genetics, including medical genetics and human genome analysis. It covers all aspects of human genetics, including molecular genetics, clinical genetics, behavioral genetics, immunogenetics, pharmacogenomics, population genetics, functional genomics, epigenetics, genetic counseling and gene therapy.
Articles on the following areas are especially welcome: genetic factors of monogenic and complex disorders, genome-wide association studies, genetic epidemiology, cancer genetics, personal genomics, genotype-phenotype relationships and genome diversity.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


