策展的癌细胞图谱提供了单细胞分辨率的肿瘤的全面表征。
IF 28.5
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
近年来,单细胞癌症研究迅速增加,但这些研究大多只分析了很少的肿瘤,这限制了它们的统计能力。将不同研究的数据和结果结合起来有很大的希望,但也涉及各种挑战。我们最近开始通过收集大量癌症单细胞rna测序数据集来解决这些挑战,利用它对肿瘤异质性进行系统分析。在这里,我们极大地扩展了这个存储库,包括超过40种癌症类型的124个数据集,总共包括2,836个样本,并改进了数据注释、可视化和探索。利用这一庞大的队列,我们生成了恶性细胞中复发表达程序的最新图谱,并系统地量化了细胞类型和癌症类型中上下文相关的基因表达和细胞周期模式。这些数据、注释和分析结果都可以通过Curated Cancer Cell Atlas免费探索和下载,这是一个为癌症研究开辟新途径的中央社区资源。本文章由计算机程序翻译,如有差异,请以英文原文为准。
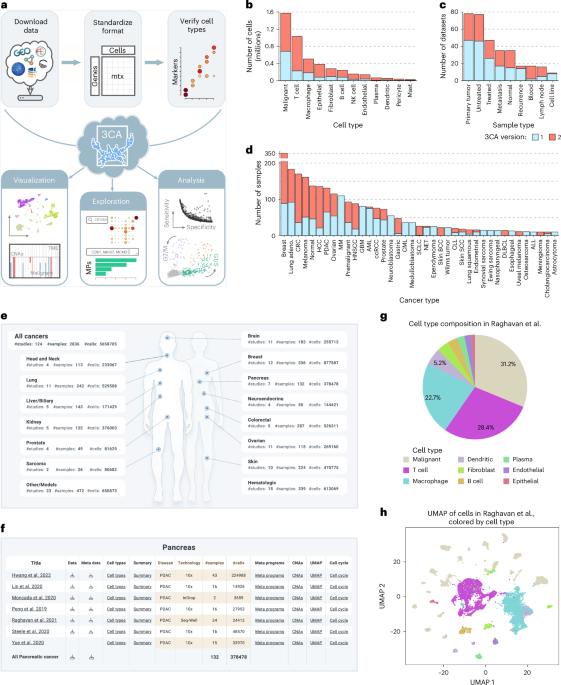
The Curated Cancer Cell Atlas provides a comprehensive characterization of tumors at single-cell resolution
Recent years have seen a rapid proliferation of single-cell cancer studies, yet most of these studies profiled few tumors, limiting their statistical power. Combining data and results across studies holds great promise but also involves various challenges. We recently began to address these challenges by curating a large collection of cancer single-cell RNA-sequencing datasets, leveraging it for systematic analyses of tumor heterogeneity. Here we greatly extend this repository to 124 datasets for over 40 cancer types, together comprising 2,836 samples, with improved data annotations, visualizations and exploration. Using this vast cohort, we generate an updated map of recurrent expression programs in malignant cells and systematically quantify context-dependent gene expression and cell-cycle patterns across cell types and cancer types. These data, annotations and analysis results are all freely available for exploration and download through the Curated Cancer Cell Atlas, a central community resource that opens new avenues in cancer research. Tyler et al. introduce a curated collection of 124 multicancer, single-cell RNA-sequencing datasets with 2,836 samples and present recurrent cancer cell expression metaprograms, quantifying context-dependent expression and proliferation across cell and cancer types.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature cancer
Medicine-Oncology
CiteScore
31.10
自引率
1.80%
发文量
129
期刊介绍:
Cancer is a devastating disease responsible for millions of deaths worldwide. However, many of these deaths could be prevented with improved prevention and treatment strategies. To achieve this, it is crucial to focus on accurate diagnosis, effective treatment methods, and understanding the socioeconomic factors that influence cancer rates.
Nature Cancer aims to serve as a unique platform for sharing the latest advancements in cancer research across various scientific fields, encompassing life sciences, physical sciences, applied sciences, and social sciences. The journal is particularly interested in fundamental research that enhances our understanding of tumor development and progression, as well as research that translates this knowledge into clinical applications through innovative diagnostic and therapeutic approaches. Additionally, Nature Cancer welcomes clinical studies that inform cancer diagnosis, treatment, and prevention, along with contributions exploring the societal impact of cancer on a global scale.
In addition to publishing original research, Nature Cancer will feature Comments, Reviews, News & Views, Features, and Correspondence that hold significant value for the diverse field of cancer research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


