定量解剖农杆菌T-DNA在单个植物细胞中的表达揭示了密度依赖性协同作用和拮抗作用
IF 13.6
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
摘要
农杆菌的发病机制涉及将T-DNA转移到植物细胞中,是植物基因工程的基石。随着依赖农杆菌的应用越来越复杂,在单个植物细胞水平上实现T-DNA表达的定量和预测性理解变得至关重要。在这里,我们研究了病原体和宿主细胞之间相互作用的经典泊松模型是否适用于农杆菌感染本烟菌。系统地挑战这个模型揭示了细菌之间不需要群体感应的拮抗和协同密度依赖相互作用。利用各种方法,我们研究了这些相互作用的分子基础。为了克服拮抗所带来的工程限制,我们创建了一个名为“BiBi”的双二进制矢量系统,它可以以预测的方式提高重组复杂代谢途径的效率。我们的发现说明了如何将理论模型与定量实验相结合可以揭示细菌发病机制的新原理,从而影响基础和应用植物生物学。本文章由计算机程序翻译,如有差异,请以英文原文为准。
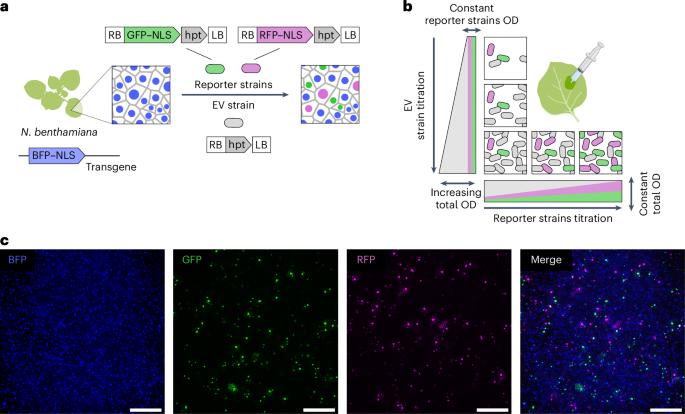

Quantitative dissection of Agrobacterium T-DNA expression in single plant cells reveals density-dependent synergy and antagonism
Agrobacterium pathogenesis, which involves transferring T-DNA into plant cells, is the cornerstone of plant genetic engineering. As the applications that rely on Agrobacterium increase in sophistication, it becomes critical to achieve a quantitative and predictive understanding of T-DNA expression at the level of single plant cells. Here we examine if a classic Poisson model of interactions between pathogens and host cells holds true for Agrobacterium infecting Nicotiana benthamiana. Systematically challenging this model revealed antagonistic and synergistic density-dependent interactions between bacteria that do not require quorum sensing. Using various approaches, we studied the molecular basis of these interactions. To overcome the engineering constraints imposed by antagonism, we created a dual binary vector system termed ‘BiBi’, which can improve the efficiency of a reconstituted complex metabolic pathway in a predictive fashion. Our findings illustrate how combining theoretical models with quantitative experiments can reveal new principles of bacterial pathogenesis, impacting both fundamental and applied plant biology. Alamos et al. show that Agrobacterium engages in density-dependent interactions that dictate transformation efficiency and transgene expression. These findings enable a quantitative model to improve metabolic engineering in a predictive manner.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


