通过连续的细胞状态分析了解风湿病
IF 32.7
1区 医学
Q1 RHEUMATOLOGY
引用次数: 0
摘要
自身免疫性风湿性疾病是一种异质性疾病,包括类风湿关节炎(RA)和系统性红斑狼疮。随着大型单细胞数据集的增加,新的疾病相关细胞类型继续在多个组学层面被识别和表征,例如RA中的“T外周辅助细胞”(TPH) (CXCR5−PD-1hi)细胞和RA中的系统性红斑狼疮和MerTK+髓样细胞。尽管人们努力定义与疾病相关的细胞图谱,但随着更高分辨率的检测方法的出现,“细胞类型”或“谱系”的定义已被证明是有争议的。本文综述了参与疾病发病机制的细胞类型和状态,重点介绍了免疫和间质细胞分类的转变观点。这些对细胞身份的理解与分析所采用的计算方法密切相关,对单细胞数据的解释有影响。了解疾病的潜在细胞结构对治疗研究也至关重要,因为模糊性阻碍了临床环境的转化。我们讨论了不同的细胞识别框架对疾病治疗的影响,以及分层药物的预测性生物标志物的发现-自身免疫性风湿性疾病未满足的临床需求。本文章由计算机程序翻译,如有差异,请以英文原文为准。
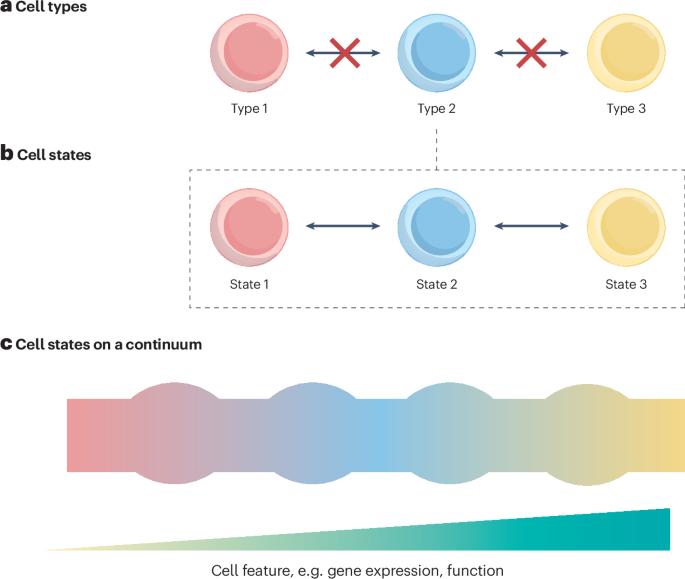

Understanding rheumatic disease through continuous cell state analysis
Autoimmune rheumatic diseases are a heterogeneous group of conditions, including rheumatoid arthritis (RA) and systemic lupus erythematosus. With the increasing availability of large single-cell datasets, novel disease-associated cell types continue to be identified and characterized at multiple omics layers, for example, ‘T peripheral helper’ (TPH) (CXCR5− PD-1hi) cells in RA and systemic lupus erythematosus and MerTK+ myeloid cells in RA. Despite efforts to define disease-relevant cell atlases, the very definition of a ‘cell type’ or ‘lineage’ has proven controversial as higher resolution assays emerge. This Review explores the cell types and states involved in disease pathogenesis, with a focus on the shifting perspectives on immune and stromal cell taxonomy. These understandings of cell identity are closely related to the computational methods adopted for analysis, with implications for the interpretation of single-cell data. Understanding the underlying cellular architecture of disease is also crucial for therapeutic research as ambiguity hinders translation to the clinical setting. We discuss the implications of different frameworks for cell identity for disease treatment and the discovery of predictive biomarkers for stratified medicine — an unmet clinical need for autoimmune rheumatic diseases. This Review explores the ongoing debate regarding the definition of cell identity, with a focus on the immune and stromal cell landscape within rheumatology. The authors discuss the implications of different frameworks of cell identity for disease treatment and the discovery of predictive biomarkers for stratified medicine.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Reviews Rheumatology
医学-风湿病学
CiteScore
29.90
自引率
0.90%
发文量
137
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Rheumatology is part of the Nature Reviews portfolio of journals. The journal scope covers the entire spectrum of rheumatology research. We ensure that our articles are accessible to the widest possible audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


