蝙蝠冠状病毒ZXC21受体结合域的结构分析
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
蝙蝠冠状病毒ZXC21和ZC45在COVID-19爆发前被发现,与SARS-CoV-2具有约86%的基因组同源性。早期研究表明,ZXC21和ZC45可能参与了SARS-CoV-2的出现。然而,ZXC21和ZC45的细胞侵袭机制尚不清楚。在这里,我们确定了ZXC21受体结合域(RBD)的晶体结构,发现其核心结构与SARS-CoV-2、MERS-CoV、人类冠状病毒(HCoV)-HKU1、SARS-CoV和HCoV- oc43的RBD具有高度相似性,而受体结合基序(RBMs)不同。我们通过表面等离子体共振(SPR)证实了ZXC21 RBD与人冠状病毒受体血管紧张素转换酶2 (ACE2)、二肽基肽酶4 (DPP4)、氨基肽酶N (APN)或跨膜丝氨酸蛋白酶2 (TMPRSS2)没有相互作用。此外,P5S-3B11 Fab可以与ZXC21 RBD结合,表明该SARS-CoV-2核心靶向抗体可能保留对ZXC21冠状病毒的中和活性。我们的研究结果揭示了蝙蝠冠状病毒ZXC21 RBD结构,这可能为SARS-CoV-2和其他人类β -冠状病毒的进化提供进一步的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
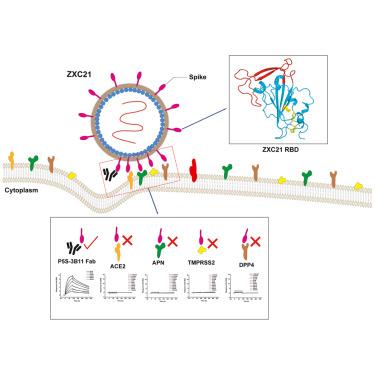
Structural insights into the receptor-binding domain of bat coronavirus ZXC21
Bat coronaviruses ZXC21 and ZC45 were discovered before the COVID-19 outbreak and share approximately 86% genome homology with SARS-CoV-2. Earlier studies indicated that ZXC21 and ZC45 may be involved in the emergence of SARS-CoV-2. However, the cell invasion mechanisms of ZXC21 and ZC45 remain unclear. Here, we determined the crystal structure of the ZXC21 receptor-binding domain (RBD) and found that the core structure shared high similarity with SARS-CoV-2, MERS-CoV, human coronavirus (HCoV)-HKU1, SARS-CoV, and HCoV-OC43 RBDs, whereas the receptor-binding motifs (RBMs) differ. We demonstrated that the ZXC21 RBD had no interaction with the human coronavirus receptors angiotensin-converting enzyme 2 (ACE2), dipeptidylpeptidase 4 (DPP4), aminopeptidase N (APN), or transmembrane serine protease 2 (TMPRSS2) by surface plasmon resonance (SPR). Moreover, the P5S-3B11 Fab can bind to the ZXC21 RBD, indicating that this SARS-CoV-2 core-targeting antibody may retain neutralizing activity toward the ZXC21 coronavirus. Our results revealed the bat coronavirus ZXC21 RBD structure, which may provide further insights into the evolution of SARS-CoV-2 and the other human beta-coronaviruses.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


