人类Argonaute2-siRNA复合物裂解RNA的机制
IF 25.9
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
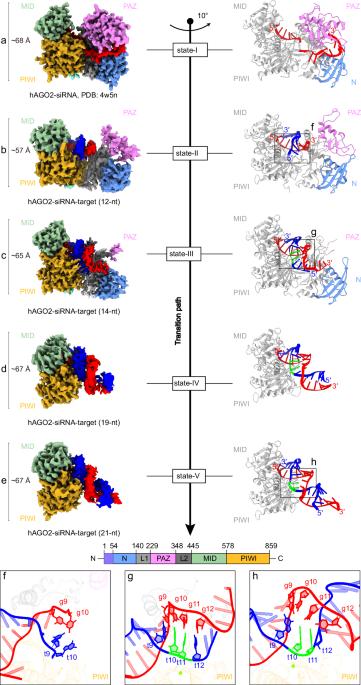
在动物实验中,AGO-clade Argonaute蛋白利用小干扰RNA (small interfering RNA, sirna)作为向导,以完全互补的方式识别靶标,从而导致靶标RNA裂解,这是实现靶标沉默的关键步骤。这些蛋白具有狭窄的核酸结合通道,限制了向导和靶之间的碱基配对超出种子区域。AGO-siRNA复合物如何克服这种结构限制并实现有效的靶标切割尚不清楚。我们使用低温电子显微镜观察与长度增加的靶rna结合的人AGO-siRNA复合物,以检查与靶识别和切割相关的构象变化。最初,构象转变从PAZ结构域的打开传播,并通过PIWI-L1-N结构域向结合通道的重新定位延伸,促进sirna -靶双工的捕获。碱基配对的后续延伸驱动PIWI-L1-N结构域向下移动,从而实现催化活化。最后,进一步在siRNA的3 '端进行碱基配对会破坏PAZ-N结构域的稳定性,导致“单叶”结构,这可能有助于AGO-siRNA酶复合物的多次翻转作用。与piwi分支Argonautes相反,AGO复合物的“单叶”结构与sirna -靶标双链的中心区域的靶标进行多次接触,将其定位在催化位点内。我们的研究结果揭示了AGO - sirna复合物执行靶RNA切割的逐步机制,并提供了AGO和PIWI蛋白在实现这种切割时不同的操作模式的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Mechanistic insights into RNA cleavage by human Argonaute2–siRNA complex
In animals, AGO-clade Argonaute proteins utilize small interfering RNAs (siRNAs) as guides to recognize target with complete complementarity, resulting in target RNA cleavage that is a critical step for target silencing. These proteins feature a constricted nucleic acid-binding channel that limits base pairing between the guide and target beyond the seed region. How the AGO–siRNA complexes overcome this structural limitation and achieve efficient target cleavage remains unclear. We performed cryo-electron microscopy of human AGO–siRNA complexes bound to target RNAs of increasing lengths to examine the conformational changes associated with target recognition and cleavage. Initially, conformational transition propagates from the opening of the PAZ domain and extends through a repositioning of the PIWI–L1–N domain toward the binding channel, facilitating the capture of siRNA–target duplex. Subsequent extension of base pairing drives the downward movement of the PIWI–L1–N domain to enable catalytic activation. Finally, further base pairing toward the 3′ end of siRNA destabilizes the PAZ–N domain, resulting in a “uni-lobed” architecture, which might facilitate the multi-turnover action of the AGO–siRNA enzyme complex. In contrast to PIWI-clade Argonautes, the “uni-lobed” structure of the AGO complex makes multiple contacts with the target in the central region of the siRNA–target duplex, positioning it within the catalytic site. Our findings shed light on the stepwise mechanisms by which the AGO–siRNA complex executes target RNA cleavage and offer insights into the distinct operational modalities of AGO and PIWI proteins in achieving such cleavage.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell Research
生物-细胞生物学
CiteScore
53.90
自引率
0.70%
发文量
2420
审稿时长
2.3 months
期刊介绍:
Cell Research (CR) is an international journal published by Springer Nature in partnership with the Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (CAS). It focuses on publishing original research articles and reviews in various areas of life sciences, particularly those related to molecular and cell biology. The journal covers a broad range of topics including cell growth, differentiation, and apoptosis; signal transduction; stem cell biology and development; chromatin, epigenetics, and transcription; RNA biology; structural and molecular biology; cancer biology and metabolism; immunity and molecular pathogenesis; molecular and cellular neuroscience; plant molecular and cell biology; and omics, system biology, and synthetic biology. CR is recognized as China's best international journal in life sciences and is part of Springer Nature's prestigious family of Molecular Cell Biology journals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


