基因组测序检测两个家族的嵌合缺失。
IF 2.5
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
摘要
三基基因组测序(GS)可用于外显子组测序无法解决致病变异的病例的遗传分析。在本文中,我们报告了两个不相关的致病性缺失家族(一个外显子覆盖基因组区域,另一个涉及单个外显子)通过GS成功鉴定。值得注意的是,在两个家族中都发现了马赛克缺失,通过使用Integrative Genomics Viewer、断点PCR、定量PCR和数字PCR分析GS数据,对其进行了仔细的详细评估。本研究强调了基于三重的GS的好处,它可以直接解释,并得到其他验证性实验方法的进一步辅助。本文章由计算机程序翻译,如有差异,请以英文原文为准。
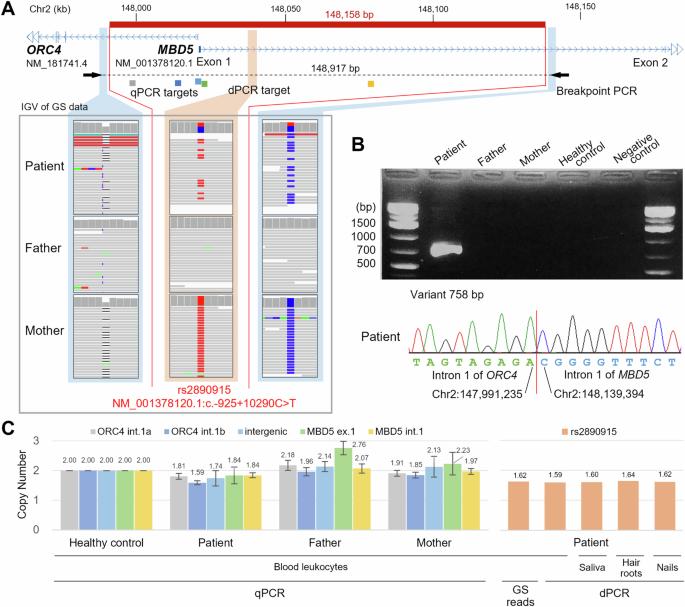
Mosaic deletions detected by genome sequencing in two families
Trio-based genome sequencing (GS) is useful for genetic analysis of cases in which exome sequencing failed to resolve the disease-causing variants. In this paper, we report two unrelated families with pathogenic deletions (one outside exome-covering genomic regions and the other involving a single exon) successfully identified by GS. Notably, mosaic deletions were found in both families, which were carefully evaluated in detail by analyzing GS data using Integrative Genomics Viewer, breakpoint PCR, quantitative PCR, and digital PCR. This study emphasizes the benefit of trio-based GS, enabling straightforward interpretation, further aided by other confirmatory experimental methods.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Human Genetics
生物-遗传学
CiteScore
7.20
自引率
0.00%
发文量
101
审稿时长
4-8 weeks
期刊介绍:
The Journal of Human Genetics is an international journal publishing articles on human genetics, including medical genetics and human genome analysis. It covers all aspects of human genetics, including molecular genetics, clinical genetics, behavioral genetics, immunogenetics, pharmacogenomics, population genetics, functional genomics, epigenetics, genetic counseling and gene therapy.
Articles on the following areas are especially welcome: genetic factors of monogenic and complex disorders, genome-wide association studies, genetic epidemiology, cancer genetics, personal genomics, genotype-phenotype relationships and genome diversity.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


