空间多组学揭示了细胞类型特异性的核室
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
哺乳动物的细胞核由不同的亚核结构划分。这些亚核结构以核体和组蛋白修饰为标志,通常是细胞类型特异性的,影响基因调控和三维基因组组织1,2,3。理解它们之间的关系取决于确定亚核结构的分子组成,并绘制它们与复杂组织中单个细胞中特定基因组位点和转录水平的关联。在这里,我们引入了双层DNA seqFISH+,它可以同时定位100,049个基因组位点,以及单个细胞中17,856个基因和亚核结构的新生转录组。这些数据使基于成像的不同亚核标记的染色质谱分析成为可能,并且可以根据标记和DNA位点,在100-200千碱基到大约1兆碱基的基因组尺度上捕获它们的变化。通过使用成年小鼠小脑的多组学数据集,我们发现抑制染色质区域比基因组中的活性区域更容易因细胞类型而变化。我们还发现,RNA聚合酶ii富集灶在局部与长细胞类型特异性基因(大于200千碱基)相关,其方式与核斑点不同。此外,我们的分析显示,由赖氨酸27位点三甲基化的组蛋白H3 (H3K27me3)和赖氨酸20位点三甲基化的组蛋白H4 (H4K20me3)标记的异染色质细胞类型特异性区域分别在特定基因和基因簇上富集,并在神经元和胶质细胞中决定径向染色体定位和染色体间相互作用。总之,我们的研究结果提供了亚核结构的单细胞高分辨率多组学视图,相关的基因组位点及其对基因调控的影响,直接在复杂组织中。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Spatial multi-omics reveals cell-type-specific nuclear compartments
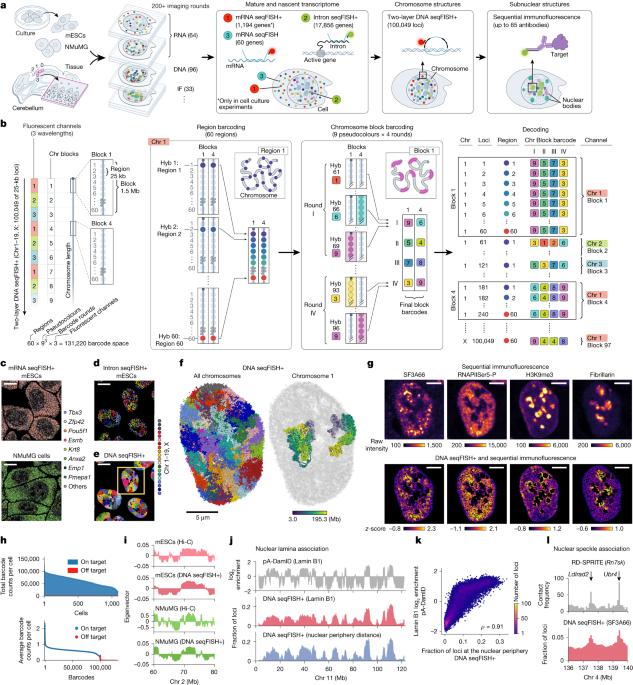
The mammalian nucleus is compartmentalized by diverse subnuclear structures. These subnuclear structures, marked by nuclear bodies and histone modifications, are often cell-type specific and affect gene regulation and 3D genome organization1–3. Understanding their relationships rests on identifying the molecular constituents of subnuclear structures and mapping their associations with specific genomic loci and transcriptional levels in individual cells, all in complex tissues. Here, we introduce two-layer DNA seqFISH+, which enables simultaneous mapping of 100,049 genomic loci, together with the nascent transcriptome for 17,856 genes and subnuclear structures in single cells. These data enable imaging-based chromatin profiling of diverse subnuclear markers and can capture their changes at genomic scales ranging from 100–200 kilobases to approximately 1 megabase, depending on the marker and DNA locus. By using multi-omics datasets in the adult mouse cerebellum, we showed that repressive chromatin regions are more variable by cell type than are active regions across the genome. We also discovered that RNA polymerase II-enriched foci were locally associated with long, cell-type-specific genes (bigger than 200 kilobases) in a manner distinct from that of nuclear speckles. Furthermore, our analysis revealed that cell-type-specific regions of heterochromatin marked by histone H3 trimethylated at lysine 27 (H3K27me3) and histone H4 trimethylated at lysine 20 (H4K20me3) are enriched at specific genes and gene clusters, respectively, and shape radial chromosomal positioning and inter-chromosomal interactions in neurons and glial cells. Together, our results provide a single-cell high-resolution multi-omics view of subnuclear structures, associated genomic loci and their effects on gene regulation, directly within complex tissues. A genomic barcoding scheme called two-layer DNA seqFISH+ enables the simultaneous mapping of more than 100,000 loci and has been used to identify cell-type-specific subnuclear compartments in the mouse brain.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


