油茶系统基因组分析及SNP指纹图谱构建。利用slf -seq技术对种质资源进行分析
IF 6.2
1区 农林科学
Q1 AGRICULTURAL ENGINEERING
引用次数: 0
摘要
油茶。山茱萸是山茱萸科的一种植物,广泛种植于中国南方,因其生产油脂和药用化合物而具有很高的价值。然而,油桐品种的规模化栽培面临着难以准确鉴定的重大挑战。针对这一问题,开展DNA指纹图谱的研究对油桐种质资源的鉴定、分类和利用具有重要意义。本研究利用特殊位点扩增片段测序(SLAF-seq)获得的SNP标记,对25个广泛栽培的油葵品种的系统发育关系和遗传多样性进行了研究。分析显示了三个不同的遗传集群,表明高度的遗传分化。值得注意的是,油桐品种CR-153表现出最高的遗传多样性,使其成为未来油桐育种计划中有希望的候选基因库。结果还表明,CL-4、DL-1和XL-1等品种;以及xml -3和xml -210,可能有共同的祖先或经历过引入事件。此外,我们还建立了包含经济性状和非生物抗性相关基因的DNA指纹图谱系统,实现了油桐的快速分类和分子育种。总之,这些发现为进一步了解中国南方栽培油桐的遗传资源提供了有价值的见解,为今后的育种规划、资源科学管理和重要经济性状的改良提供了实用指导。本文章由计算机程序翻译,如有差异,请以英文原文为准。
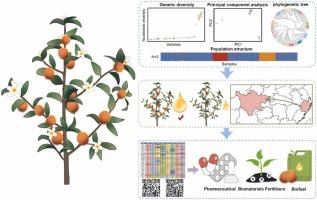
Phylogenomic analysis and SNP fingerprinting construction of Camellia oleifera Abel. germplasm resources using SLAF-seq technology
Camellia oleifera Abel., a species of Theaceae plants widely cultivated in southern China, is highly valued for its production of oils and pharmaceutical compounds. However, the large-scale cultivation of C. oleifera cultivars faces significant challenges due to difficulties in accurate identification. To address this, developing DNA fingerprinting is crucial for identifying superior germplasm and enhancing the classification and utilization of C. oleifera resources. In this study, we utilized SNP markers derived from specific-locus amplified fragment sequencing (SLAF-seq) to investigate the phylogenetic relationships and genetic diversity among 25 widely cultivated C. oleifera cultivars. The analysis revealed three distinct genetic clusters, indicating high levels of genetic differentiation. Notably, the C. oleifera cultivar CR-153 exhibited the highest genetic diversity, making it a promising candidate genetic pool for future breeding programs aimed at improving C. oleifera. The results also suggest that cultivars such as CL-4, DL-1 and XL-1; as well as XL-3 and XL-210, may share ancestry or have undergone introduction events. Furthermore, we developed a DNA fingerprinting system incorporating genes relevant to economic traits and abiotic resistance, enabling rapid classification and molecular breeding of C. oleifera. Overall, these findings provide valuable insights into the genetic resources of cultivated C. oleifera in southern China, offering practical guidelines for future breeding programs, scientific resource management, and the enhancement of economically important traits.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Industrial Crops and Products
农林科学-农业工程
CiteScore
9.50
自引率
8.50%
发文量
1518
审稿时长
43 days
期刊介绍:
Industrial Crops and Products is an International Journal publishing academic and industrial research on industrial (defined as non-food/non-feed) crops and products. Papers concern both crop-oriented and bio-based materials from crops-oriented research, and should be of interest to an international audience, hypothesis driven, and where comparisons are made statistics performed.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


