高通量单分子显微镜与适应性空间分辨率使用可交换的寡核苷酸标签
IF 16
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
摘要
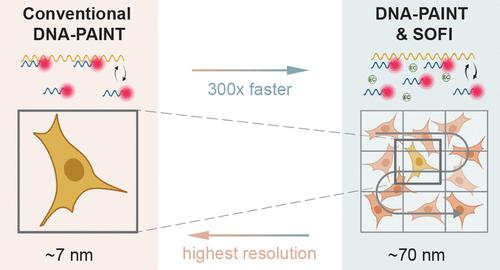
超分辨率显微镜有助于以接近分子水平的分辨率观察细胞结构。特别是基于单分子定位的超分辨率技术对仪器的要求相对较低,因此是生物成像技术的理想选择。然而,它们的低通量特性阻碍了它们在生物分子研究和筛选中的应用。在这里,我们提出了一种更高效的数据收集工作流程,首先使用基于快速波动的成像技术对大面积区域进行扫描,然后对选定的细胞进行单分子定位显微镜检查。为了实现这一工作流程,我们利用 DNA-PAINT 探针与 DNA 寡聚杂交动力学的多功能性,对荧光闪烁进行定制,以实现高通量和高分辨率成像。此外,我们还利用超分辨率光学波动成像(SOFI)来分析 DNA-PAINT 结合动力学的统计波动,从而容许更密集的闪烁并加快成像速度。因此,与传统的 DNA-PAINT 成像相比,我们对不同细胞结构的成像速度提高了 30-300 倍,尽管分辨率较低。值得注意的是,通过调整图像介质和数据处理,我们可以在高通量 SOFI(在 4 分钟的总采集时间内扫描 0.65 mm × 0.52 mm 的 FOV)和超分辨率 DNA-PAINT 显微镜之间灵活切换,从而证明 DNA-PAINT 和 SOFI 的结合可以根据成像需要调整图像分辨率和采集时间。我们设想这种方法在与多路复用和三维成像相结合时将特别强大。本文章由计算机程序翻译,如有差异,请以英文原文为准。
High-Throughput Single-Molecule Microscopy with Adaptable Spatial Resolution Using Exchangeable Oligonucleotide Labels
Super-resolution microscopy facilitates the visualization of cellular structures at a resolution approaching the molecular level. Especially, super-resolution techniques based on the localization of single molecules have relatively modest instrument requirements and are thus good candidates for adoption in bioimaging. However, their low-throughput nature hampers their applicability in biomolecular research and screening. Here, we propose a workflow for more efficient data collection, starting with the scanning of large areas using fast fluctuation-based imaging, followed by single-molecule localization microscopy of selected cells. To achieve this workflow, we exploit the versatility of DNA oligo hybridization kinetics with DNA-PAINT probes to tailor the fluorescent blinking toward high-throughput and high-resolution imaging. Additionally, we employ super-resolution optical fluctuation imaging (SOFI) to analyze statistical fluctuations in the DNA-PAINT binding kinetics, thereby tolerating much denser blinking and facilitating accelerated imaging speeds. Thus, we demonstrate 30–300-fold faster imaging of different cellular structures compared to conventional DNA-PAINT imaging, albeit at a lower resolution. Notably, by tuning the image medium and data processing though, we can flexibly switch between high-throughput SOFI (scanning an FOV of 0.65 mm × 0.52 mm within 4 min of total acquisition time) and super-resolution DNA-PAINT microscopy and thereby demonstrate that combining DNA-PAINT and SOFI enables one to adapt image resolution and acquisition time based on the imaging needs. We envision this approach to be especially powerful when combined with multiplexing and 3D imaging.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


