SERS- atb:抗生素SERS光谱可视化和深度学习识别的综合数据库服务器
IF 7.3
2区 环境科学与生态学
Q1 ENVIRONMENTAL SCIENCES
引用次数: 0
摘要
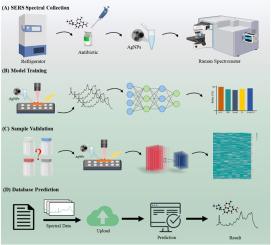
快速准确地鉴定环境样品中的抗生素对于解决日益严重的抗生素污染问题至关重要,特别是在水源中。抗生素污染通过促进抗生素耐药性的传播,对生态系统和人类健康构成重大风险。SERS以其高灵敏度和特异性而闻名,是抗生素鉴定的有力工具。然而,由于缺乏对环境和临床应用至关重要的大规模抗生素光谱数据库,其广泛应用受到限制。为了满足这一需求,我们系统地收集了200种环境相关抗生素的12,800个SERS光谱,并在http://sers.test.bniu.net/上开发了一个开放获取的基于网络的数据库。我们将六种机器学习算法与CNN模型进行了比较,CNN模型的准确率最高,达到98.94%,使其成为首选的数据库模型。对于外部验证,CNN的准确率为82.8%,强调了其在实际应用中的可靠性和实用性。SERS数据库和CNN预测模型代表了一种新的环境监测资源,在可访问性、速度和可扩展性方面具有显著优势。本研究建立了大规模的公共抗生素SERS光谱数据库,促进了SERS与环境计划的整合,具有改善抗生素检测、污染管理和耐药性缓解的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
SERS-ATB: A comprehensive database server for antibiotic SERS spectral visualization and deep-learning identification
The rapid and accurate identification of antibiotics in environmental samples is critical for addressing the growing concern of antibiotic pollution, particularly in water sources. Antibiotic contamination poses a significant risk to ecosystems and human health by contributing to the spread of antibiotic resistance. Surface-enhanced Raman spectroscopy (SERS), known for its high sensitivity and specificity, is a powerful tool for antibiotic identification. However, its broader application is constrained by the lack of a large-scale antibiotic spectral database crucial for environmental and clinical use. To address this need, we systematically collected 12,800 SERS spectra for 200 environmentally relevant antibiotics and developed an open-access, web-based database at http://sers.test.bniu.net/. We compared six machine learning algorithms with a convolutional neural network (CNN) model, which achieved the highest accuracy at 98.94%, making it the preferred database model. For external validation, CNN demonstrated an accuracy of 82.8%, underscoring its reliability and practicality for real-world applications. The SERS database and CNN prediction model represent a novel resource for environmental monitoring, offering significant advantages in terms of accessibility, speed, and scalability. This study establishes the large-scale, public SERS spectral databases for antibiotics, facilitating the integration of SERS into environmental programs, with the potential to improve antibiotic detection, pollution management, and resistance mitigation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Environmental Pollution
环境科学-环境科学
CiteScore
16.00
自引率
6.70%
发文量
2082
审稿时长
2.9 months
期刊介绍:
Environmental Pollution is an international peer-reviewed journal that publishes high-quality research papers and review articles covering all aspects of environmental pollution and its impacts on ecosystems and human health.
Subject areas include, but are not limited to:
• Sources and occurrences of pollutants that are clearly defined and measured in environmental compartments, food and food-related items, and human bodies;
• Interlinks between contaminant exposure and biological, ecological, and human health effects, including those of climate change;
• Contaminants of emerging concerns (including but not limited to antibiotic resistant microorganisms or genes, microplastics/nanoplastics, electronic wastes, light, and noise) and/or their biological, ecological, or human health effects;
• Laboratory and field studies on the remediation/mitigation of environmental pollution via new techniques and with clear links to biological, ecological, or human health effects;
• Modeling of pollution processes, patterns, or trends that is of clear environmental and/or human health interest;
• New techniques that measure and examine environmental occurrences, transport, behavior, and effects of pollutants within the environment or the laboratory, provided that they can be clearly used to address problems within regional or global environmental compartments.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


