四纳米孔阵列使四单链DNA均聚物的高分辨率鉴定
IF 16
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
摘要
固态纳米孔技术具有开发机械稳定和小型化DNA测序设备的潜力。然而,由于纳米孔内部的高电场和缺乏有效的速度控制策略,导致时间分辨率有限,阻碍了测序的实现。在这里,我们报道了一个四阵列(四个纳米孔,孔间间距为~ 30 nm作为检测单元),该阵列诱导了纳米孔阵列内外电场的重新分布,并提供了四种ssDNA均聚物类型的高分辨率识别。我们证明了四纳米孔阵列很好地解决了polyA25的易位事件,并且具有20 nt的长度分辨率。分子动力学模拟证实了四纳米孔阵列的易位减慢和优越的性能。我们发现纳米孔阵列结构导致纳米孔内部电场的直接减小以及由于电场串扰导致纳米孔外部电场的增加。这种双重好处不仅减少了对DNA的巨大驱动力,而且促进了分子通过纳米孔捕获,从而降低了电压阈值。最后,在低至30 mV的电压和8 μs/nt的易位速度下,成功地识别了4种ssDNA均聚物(polyA25、polyT25、polyC25和polyG10)。这些发现为纳米孔阵列鉴别短单链核苷酸提供了高分辨率的见解,并展示了开发利用纳米孔阵列作为传感单元的DNA测序仪的巨大潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
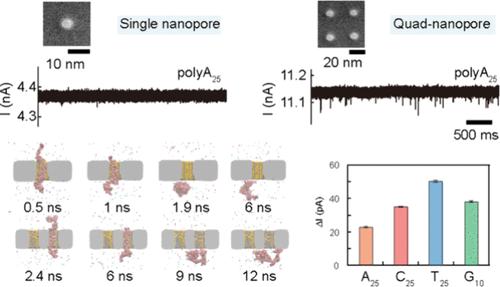
Quad-Nanopore Array Enables High-Resolution Identification of Four Single-Stranded DNA Homopolymers
The solid-state nanopore technique holds the potential to develop mechanically stable and miniaturized DNA sequencing devices. However, the limited temporal resolution due to the high electric field inside the nanopore and the lack of an effective speed control strategy have hindered the realization of sequencing. Here, we reported a quad-array (four nanopores milled with ∼30 nm interpore spacing as a detection unit) that induced a redistribution of the electric field inside and outside the nanopore array and offered high-resolution discrimination of four ssDNA homopolymer types. We demonstrated that the quad-nanopore array well resolved the translocation events of polyA25 and had a length resolution of 20 nt. The molecular dynamic simulation confirmed the slowed-down translocations and superior performance of a quad-nanopore array. We found that the nanopore array configuration induced a direct reduction of the electric field inside the nanopore as well as an increase in the electrical field outside the nanopore due to electric field crosstalk. This dual benefit not only reduced the large driving force on DNA but also facilitated molecule capture through nanopores, therefore decreasing the voltage thresholds. Finally, the successful discrimination of four ssDNA homopolymer types (polyA25, polyT25, polyC25, and polyG10) was achieved using a voltage as low as 30 mV with a translocation speed of 8 μs/nt. These findings provide insights into nanopore arrays for discriminating short single-stranded nucleotides with high resolution and demonstrate promising potential for developing DNA sequencers that utilize nanopore arrays as sensing units.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


