水稻根系内生微生物群中关键类群及其相关噬菌体的群落结构和代谢潜力对金属污染的响应
IF 7.3
2区 环境科学与生态学
Q1 ENVIRONMENTAL SCIENCES
引用次数: 0
摘要
农产品重金属污染直接威胁食品安全,已成为全球关注的环境问题。植物相关微生物群,特别是内生微生物群,具有缓解HM胁迫和促进植物生长的潜力。内生菌的代谢潜力,特别是在HM胁迫下的代谢潜力尚未得到很好的解决。水稻是世界主要的主食,与其他作物相比,它更容易受到HM污染,因此需要特别关注。因此,本研究选择水稻作为目标植物。采用地球化学分析和扩增子测序相结合的方法,对2个hm污染稻田的水稻根系内生细菌群落进行了分析,并鉴定了重点分类群。利用宏基因组分析对这些关键类群的代谢潜力进行了研究。鉴定出Burkholderiales和Rhizobiales为优势楔状分类群。与这些关键群体相关的宏基因组组装基因组(MAG)表明,它们具有与金属抗性和转化(例如,As抗性和循环,V还原,Cr外排和还原)以及植物生长促进(固氮,磷酸盐增溶,氧化胁迫抗性,吲哚-3-乙酸和铁载体生产)相关的多种遗传潜力。此外,还发现了编码与HM抗性以及氮和磷酸盐获取相关的辅助代谢基因(AMGs)的噬菌体,这表明这些噬菌体可能有助于水稻根系中这些关键的生物地球化学过程。本研究结果揭示了水稻内生关键类群及其相关噬菌体在hm污染水稻根系内生微生物组中的有益作用,为今后利用根系微生物组进行农业生产安全管理提供了有价值的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
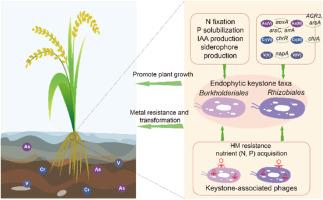
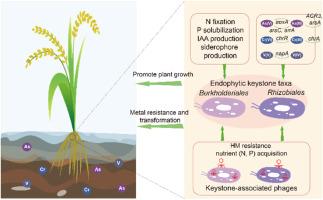
Community structure and metabolic potentials of keystone taxa and their associated bacteriophages within rice root endophytic microbiome in response to metal(loid)s contamination
Heavy metal (HM) contamination of agricultural products is of global environmental concern as it directly threatened the food safety. Plant-associated microbiome, particularly endophytic microbiome, hold the potential for mitigating HM stress as well as promoting plant growth. The metabolic potentials of the endophytes, especially those under the HM stresses, have not been well addressed. Rice, a major staple food worldwide, is more vulnerable to HM contamination compared to other crops and therefore requires special attentions. Therefore, this study selected rice as the target plants. Geochemical analysis and amplicon sequencing were combined to characterize the rice root endophytic bacterial communities and identify keystone taxa in two HM-contaminated rice fields. Metagenomic analysis was employed to investigate the metabolic potentials of these keystone taxa. Burkholderiales and Rhizobiales were identified as predominant keystone taxa. The metagenome-assembled genome (MAG)s associated with these keystone populations suggested that they possessed diverse genetic potentials related to metal resistance and transformation (e.g., As resistance and cycling, V reduction, Cr efflux and reduction), and plant growth promotion (nitrogen fixation, phosphate solubilization, oxidative stress resistance, indole-3-acetic acid, and siderophore production). Moreover, bacteriophages encoding auxiliary metabolism genes (AMGs) associated with the HM resistance as well as nitrogen and phosphate acquisition were identified, suggesting that these phages may contribute to these crucial biogeochemical processes within rice roots. The current findings revealed the beneficial roles of rice endophytic keystone taxa and their associated bacteriophages within HM-contaminated rice root endophytic microbiome, which may provide valuable insights on future applications of employing root microbiome for safety management of agriculture productions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Environmental Pollution
环境科学-环境科学
CiteScore
16.00
自引率
6.70%
发文量
2082
审稿时长
2.9 months
期刊介绍:
Environmental Pollution is an international peer-reviewed journal that publishes high-quality research papers and review articles covering all aspects of environmental pollution and its impacts on ecosystems and human health.
Subject areas include, but are not limited to:
• Sources and occurrences of pollutants that are clearly defined and measured in environmental compartments, food and food-related items, and human bodies;
• Interlinks between contaminant exposure and biological, ecological, and human health effects, including those of climate change;
• Contaminants of emerging concerns (including but not limited to antibiotic resistant microorganisms or genes, microplastics/nanoplastics, electronic wastes, light, and noise) and/or their biological, ecological, or human health effects;
• Laboratory and field studies on the remediation/mitigation of environmental pollution via new techniques and with clear links to biological, ecological, or human health effects;
• Modeling of pollution processes, patterns, or trends that is of clear environmental and/or human health interest;
• New techniques that measure and examine environmental occurrences, transport, behavior, and effects of pollutants within the environment or the laboratory, provided that they can be clearly used to address problems within regional or global environmental compartments.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


