通过对t细胞空间蛋白质组谱的反事实学习,识别促进t细胞浸润肿瘤的扰动
IF 26.8
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
摘要
通过激活内源性或工程化T细胞及其对肿瘤微环境的浸润,可以减缓或停止癌症的进展。在这里,我们描述了一个深度学习模型,该模型使用肿瘤的大规模空间蛋白质组学谱来产生促进t细胞浸润的最小肿瘤扰动。该模型将扰动生成的反事实优化策略与t细胞浸润的预测集成为一个自监督机器学习问题。我们将该模型应用于368例转移性黑色素瘤和结直肠癌样本,使用40层成像细胞计数仪进行检测,并发现队列依赖的组合扰动(黑色素瘤的CXCL9, CXCL10, CCL22和CCL18,结直肠癌的CXCR4, PD-1, PD-L1和CYR61)支持t细胞在患者队列中的浸润,通过体外实验证实了这一点。利用基于反事实的空间组学数据预测可能有助于癌症治疗方法的设计。本文章由计算机程序翻译,如有差异,请以英文原文为准。

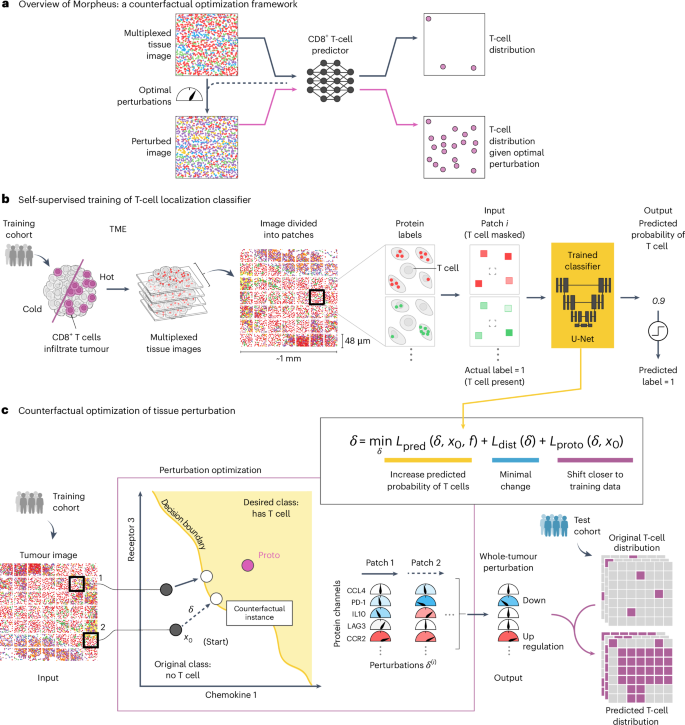
Identifying perturbations that boost T-cell infiltration into tumours via counterfactual learning of their spatial proteomic profiles
Cancer progression can be slowed down or halted via the activation of either endogenous or engineered T cells and their infiltration of the tumour microenvironment. Here we describe a deep-learning model that uses large-scale spatial proteomic profiles of tumours to generate minimal tumour perturbations that boost T-cell infiltration. The model integrates a counterfactual optimization strategy for the generation of the perturbations with the prediction of T-cell infiltration as a self-supervised machine learning problem. We applied the model to 368 samples of metastatic melanoma and colorectal cancer assayed using 40-plex imaging mass cytometry, and discovered cohort-dependent combinatorial perturbations (CXCL9, CXCL10, CCL22 and CCL18 for melanoma, and CXCR4, PD-1, PD-L1 and CYR61 for colorectal cancer) that support T-cell infiltration across patient cohorts, as confirmed via in vitro experiments. Leveraging counterfactual-based predictions of spatial omics data may aid the design of cancer therapeutics. Minimal tumour perturbations that boost T-cell infiltration can be discovered by using deep learning to analyse large-scale spatial omics profiles of tumours.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Biomedical Engineering
Medicine-Medicine (miscellaneous)
CiteScore
45.30
自引率
1.10%
发文量
138
期刊介绍:
Nature Biomedical Engineering is an online-only monthly journal that was launched in January 2017. It aims to publish original research, reviews, and commentary focusing on applied biomedicine and health technology. The journal targets a diverse audience, including life scientists who are involved in developing experimental or computational systems and methods to enhance our understanding of human physiology. It also covers biomedical researchers and engineers who are engaged in designing or optimizing therapies, assays, devices, or procedures for diagnosing or treating diseases. Additionally, clinicians, who make use of research outputs to evaluate patient health or administer therapy in various clinical settings and healthcare contexts, are also part of the target audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


