用于早期癌症检测的无细胞 DNA 基因组和片段组图谱
IF 66.8
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
血浆中游离细胞DNA (cfDNA)的基因组分析使无创血液生物标志物方法能够用于癌症检测和疾病监测。目前鉴定循环肿瘤DNA的方法通常使用靶向肿瘤特异性突变或甲基化分析。一种新兴的方法是基于对癌症患者全基因组cfDNA片段改变的识别。最近的研究揭示了许多可以影响基因组中cfDNA片段汇编的特征,这些特征统称为“cfDNA片段组”。这些变化源于个体的基因组、表观基因组、转录组和染色质状态,并影响cfDNA的大小、位置、覆盖范围、突变、结构和甲基化特征。识别和监测这些变化有可能改善癌症的早期发现,特别是使用高度敏感的多特征机器学习方法,这将适用于风险增加的人群的广泛使用。本文重点介绍了快速发展的cfDNA全基因组特征分析领域,与现有cfDNA方法的比较,以及最近在大规模测序和人工智能交叉领域的相关创新。由于cfDNA片段组方法的广泛临床应用对癌症筛查和癌症患者临床管理的个性化方法具有巨大的公共卫生影响,我们概述了未来的挑战和机遇。本文章由计算机程序翻译,如有差异,请以英文原文为准。
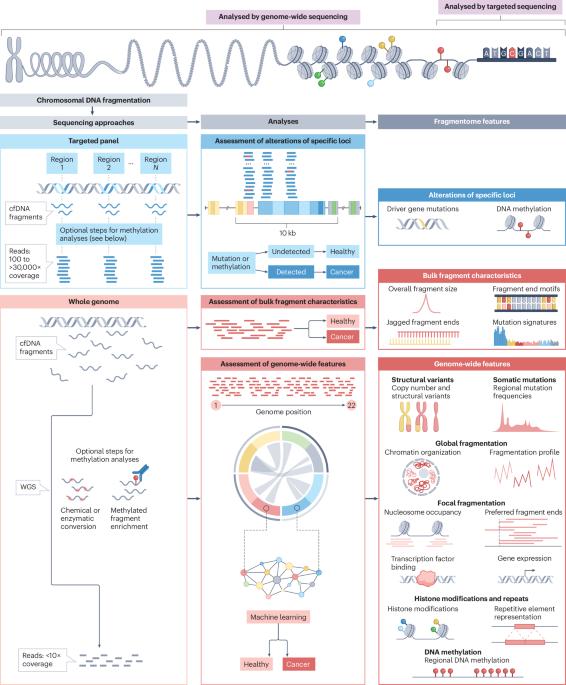
Genomic and fragmentomic landscapes of cell-free DNA for early cancer detection
Genomic analyses of cell-free DNA (cfDNA) in plasma are enabling noninvasive blood-based biomarker approaches to cancer detection and disease monitoring. Current approaches for identification of circulating tumour DNA typically use targeted tumour-specific mutations or methylation analyses. An emerging approach is based on the recognition of altered genome-wide cfDNA fragmentation in patients with cancer. Recent studies have revealed a multitude of characteristics that can affect the compendium of cfDNA fragments across the genome, collectively called the ‘cfDNA fragmentome’. These changes result from genomic, epigenomic, transcriptomic and chromatin states of an individual and affect the size, position, coverage, mutation, structural and methylation characteristics of cfDNA. Identifying and monitoring these changes has the potential to improve early detection of cancer, especially using highly sensitive multi-feature machine learning approaches that would be amenable to broad use in populations at increased risk. This Review highlights the rapidly evolving field of genome-wide analyses of cfDNA characteristics, their comparison to existing cfDNA methods, and recent related innovations at the intersection of large-scale sequencing and artificial intelligence. As the breadth of clinical applications of cfDNA fragmentome methods have enormous public health implications for cancer screening and personalized approaches for clinical management of patients with cancer, we outline the challenges and opportunities ahead. In this Review, Bruhm et al. provide a comprehensive overview of targeted and genome-wide cell-free DNA detection approaches, examining their potential to complement existing screening programmes or enhance early cancer detection for those cancers without effective screening methods.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Reviews Cancer
医学-肿瘤学
CiteScore
111.90
自引率
0.40%
发文量
97
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Cancer, a part of the Nature Reviews portfolio of journals, aims to be the premier source of reviews and commentaries for the scientific communities it serves. The correct abbreviation for abstracting and indexing purposes is Nat. Rev. Cancer. The international standard serial numbers (ISSN) for Nature Reviews Cancer are 1474-175X (print) and 1474-1768 (online). Unlike other journals, Nature Reviews Cancer does not have an external editorial board. Instead, all editorial decisions are made by a team of full-time professional editors who are PhD-level scientists. The journal publishes Research Highlights, Comments, Reviews, and Perspectives relevant to cancer researchers, ensuring that the articles reach the widest possible audience due to their broad scope.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


