常见癌症的转录组结构反映了合成的致死相互作用
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
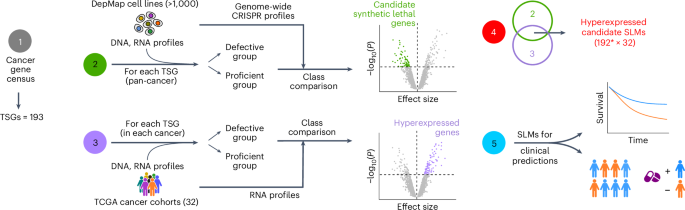
为了维持细胞的适应性,有害的基因改变被其他基因的代偿性变化所缓冲。在癌症中,缓冲过程可以被合成致死性靶向。然而,尽管在临床前模型中大规模确定了合成致死效应,但证明这些效应在临床上起作用的证据有限。这阻碍了合成致死方法的应用。通过整合来自9000例癌症和合成致死筛选的分子谱数据,我们发现,通过合成致死伴侣的高表达来抑制肿瘤抑制基因(TSG)丢失的转录组缓冲是一种普遍现象,并扩展到多种TSG和组织类型。转录组缓冲在TSG表型缺失的癌症中也很明显,如BRCAness癌症,BRCA1/2合成致死基因的表达与临床结果相关。表现出转录组缓冲的合成致死基因也表现出更强的合成致死效应。这些观察结果有助于理解肿瘤细胞如何耐受TSG丢失,部分解释癌症中的转录组结构,并为靶标选择提供见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。

The transcriptomic architecture of common cancers reflects synthetic lethal interactions
To maintain cell fitness, deleterious genetic alterations are buffered by compensatory changes in additional genes. In cancer, buffering processes could be targeted by synthetic lethality. However, despite the large-scale identification of synthetic lethal effects in preclinical models, evidence that these operate clinically is limited. This impedes the application of synthetic lethal approaches. By integrating molecular profiling data from >9,000 cancers with synthetic lethal screens, we show that transcriptomic buffering of tumor suppressor gene (TSG) loss by hyperexpression of synthetic lethal partners is a common phenomenon, extending to multiple TSGs and histotypes. Transcriptomic buffering is also notable in cancers that phenocopy TSG loss, such as BRCAness cancers, where expression of BRCA1/2 synthetic lethal genes correlates with clinical outcome. Synthetic lethal genes that exhibit transcriptomic buffering also represent more robust synthetic lethal effects. These observations have implications for understanding how tumor cells tolerate TSG loss, in part explain transcriptomic architectures in cancer and provide insight into target selection. Tumor cells upregulate compensatory buffering genes following tumor suppressor loss. These genes may represent new synthetic lethal partners that could be harnessed therapeutically.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


