多组学分析发现参与 Codonopsis pilosula (Franch.) Nannf.中三萜类和倍半萜生物合成的新候选基因
IF 6.2
1区 农林科学
Q1 AGRICULTURAL ENGINEERING
引用次数: 0
摘要
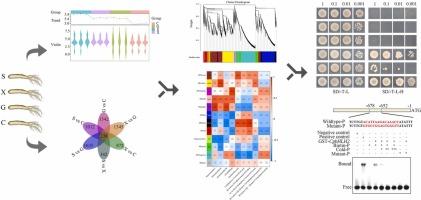
党参是生物活性倍半萜和三萜的重要来源。尽管如此,控制倍半萜类和三萜类生物合成的调节网络在党参中仍然主要是未知的。在这项研究中,我们进行了广泛的代谢组学和转录组学研究,以揭示基因网络,确定关键的候选基因,以及控制三萜类和倍半萜类物质产生的转录因子。我们的研究发现了15种不同的三萜类和倍半萜类,其中10种基于它们的起源表现出不同的积累。利用Mfuzz聚类和加权基因共表达网络分析,我们分离出代谢物特异性模块/簇。共鉴定出46个可能参与三萜类和倍半萜类合成的基因和转录因子,其中20个基因与萜类骨架合成相关,26个基因与三萜类和倍半萜类的产生和调节相关。进一步的酵母单杂交和电泳迁移转移实验表明,CpbHLH2、CpbHLH3和CpAP2/ERF2可能通过直接调控CpLUP4-14、CpSQLE-2和CpFDFT1-8的转录而显著影响三萜和倍半萜的生物合成。综上所述,本研究结果为深入了解三萜类和倍半萜类生物合成的分子机制提供了有价值的见解,为进一步研究三萜类和倍半萜类化合物在党参根中的积累提供了遗传资源和指导。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Multi-omic analysis highlights new candidate genes involved in triterpenoid and sesquiterpenoid biosynthesis in Codonopsis pilosula (Franch.) Nannf.
Codonopsis pilosula root is a notable resource for bioactive sesquiterpenoids and triterpenoids. Despite this, the regulatory networks governing sesquiterpenoid and triterpenoid biosynthesis within C. pilosula, remain predominantly uncharted. In this study, we performed extensive metabolome and transcriptome investigations to reveal the gene networks, pinpoint crucial candidate genes, and transcription factors that govern the production of triterpenoids and sesquiterpenoids in C. pilosula. Our research characterized 15 distinct triterpenoids and sesquiterpenoids, with 10 showing differential accumulation based on their origins. Utilizing Mfuzz clustering and weighted gene co-expression network analysis, we isolated metabolite-specific modules/clusters. A total of 46 genes and some transcription factors potentially involved in triterpenoid and sesquiterpenoid production were identified, comprising 20 genes linked to terpenoid skeleton synthesis and 26 genes tied to the generation and modulation of triterpenoids and sesquiterpenoids. Further assays, including yeast one-hybrid and electrophoretic mobility shift, indicated that CpbHLH2, CpbHLH3, and CpAP2/ERF2 may significantly influence triterpenoid and sesquiterpenoid biosynthesis by directly regulating the transcription of CpLUP4–14, CpSQLE-2, and CpFDFT1–8. In conclusion, our results provide valuable insights into the molecular mechanisms controlling triterpenoid and sesquiterpenoid biosynthesis in C. pilosula, enhancing genetic resources and guiding future research on these compounds’ accumulation in C. pilosula root.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Industrial Crops and Products
农林科学-农业工程
CiteScore
9.50
自引率
8.50%
发文量
1518
审稿时长
43 days
期刊介绍:
Industrial Crops and Products is an International Journal publishing academic and industrial research on industrial (defined as non-food/non-feed) crops and products. Papers concern both crop-oriented and bio-based materials from crops-oriented research, and should be of interest to an international audience, hypothesis driven, and where comparisons are made statistics performed.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


