转录适应在杜氏肌营养不良中上调营养蛋白
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
杜氏肌营养不良症(DMD)是一种由编码肌营养不良蛋白1,2的DMD基因突变引起的肌肉退行性疾病。肌营养蛋白(UTRN)是DMD的遗传和功能同源蛋白,在一些DMD患者中表达上调3,4,5。为了进一步研究这种UTRN上调,我们首先通过在其可选剪接的外显子之一中引入过早终止密码子(PTC),开发了DMD的诱导信使RNA (mRNA)降解系统。包含ptc的外显子触发DMD突变体mRNA衰变和UTRN上调。值得注意的是,阻断无义介导的mRNA衰减会导致UTRN上调的逆转,而过表达DMD则不会。此外,在野生型细胞中过表达DMDPTC小基因会导致UTRN上调,野生型DMD小基因含有自裂核酶也是如此。为了将这些发现置于治疗背景下,我们使用剪接开关反义寡核苷酸(ASOs)诱导DMD外框外显子的跳跃,旨在引入ptc。我们发现这些aso会导致UTRN上调。此外,当使用ASO恢复来自DMDΔE52患者的肌管的DMD阅读框时,UTRN上调减少了。总之,这些结果表明,一种基于mRNA衰变的转录适应机制6,7,8在DMDPTC患者的UTRN上调中起着关键作用,并且它们突出了ASOs和核酶在通过转录适应诱导遗传补偿方面尚未被探索的治疗应用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
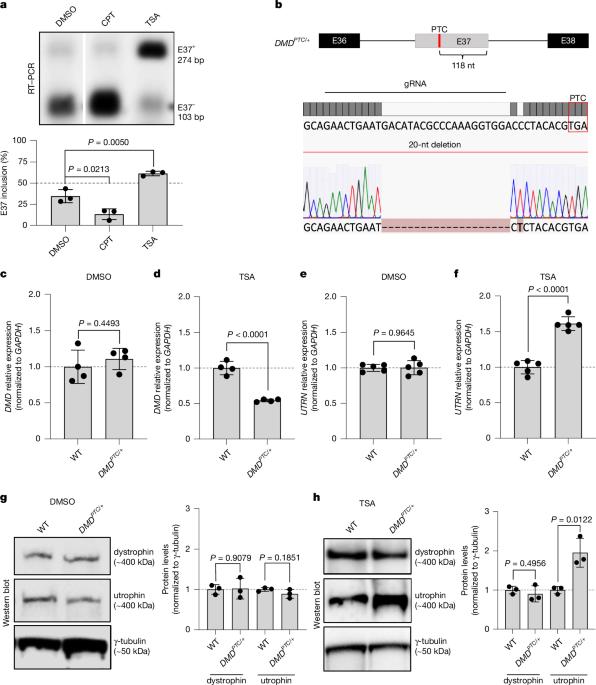
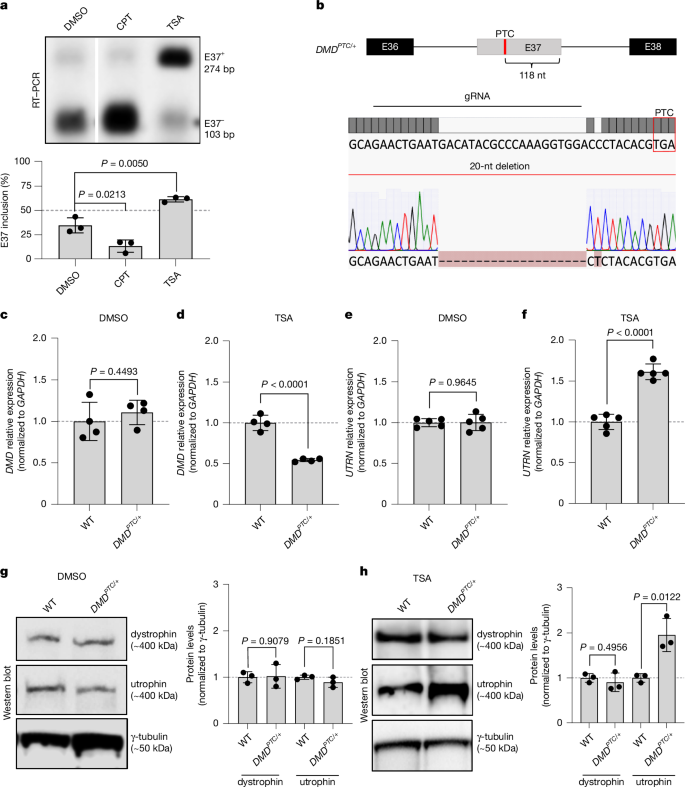
Transcriptional adaptation upregulates utrophin in Duchenne muscular dystrophy
Duchenne muscular dystrophy (DMD) is a muscle-degenerating disease caused by mutations in the DMD gene, which encodes the dystrophin protein1,2. Utrophin (UTRN), the genetic and functional paralogue of DMD, is upregulated in some DMD patients3–5. To further investigate this UTRN upregulation, we first developed an inducible messenger RNA (mRNA) degradation system for DMD by introducing a premature termination codon (PTC) in one of its alternatively spliced exons. Inclusion of the PTC-containing exon triggers DMD mutant mRNA decay and UTRN upregulation. Notably, blocking nonsense-mediated mRNA decay results in the reversal of UTRN upregulation, whereas overexpressing DMD does not. Furthermore, overexpressing DMDPTC minigenes in wild-type cells causes UTRN upregulation, as does a wild-type DMD minigene containing a self-cleaving ribozyme. To place these findings in a therapeutic context, we used splice-switching antisense oligonucleotides (ASOs) to induce the skipping of out-of-frame exons of DMD, aiming to introduce PTCs. We found that these ASOs cause UTRN upregulation. In addition, when using an ASO to restore the DMD reading frame in myotubes derived from a DMDΔE52 patient, an actual DMD treatment, UTRN upregulation was reduced. Altogether, these results indicate that an mRNA decay-based mechanism called transcriptional adaptation6–8 plays a key role in UTRN upregulation in DMDPTC patients, and they highlight an unexplored therapeutic application of ASOs, as well as ribozymes, in inducing genetic compensation via transcriptional adaptation. Transcriptional adaptation upregulates UTRN in Duchenne muscular dystrophy (DMD) patients, as supported by several lines of evidence, including the use of splice-switching antisense oligonucleotides to induce the skipping of out-of-frame exons of the DMD gene.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


