IF 5.7
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
背景可靠的组织预处理和脂质提取工作流程对于代谢表型研究和准确解读脂质图谱至关重要。目前已开发出多种组织脂质提取方法,但技术的选择会影响分析结果。本研究对用于液相色谱-质谱法脂质组分析的六种液-液萃取方法(三种双相法和三种单相法)进行了全面评估。评估了这些萃取方法是否适用于对脂肪、肝脏和心脏等不同类型的组织进行全面的脂质分析。研究结果表明,根据组织类型的不同,使用每种技术提取的脂质种类在覆盖范围和可靠性方面存在显著差异。脂肪组织的最佳提取方法是丁醇:甲醇(BUME)(3:1),它的脂质覆盖率、产量和重现性最高(886 个脂质,变异系数为 30%);甲基叔丁基醚(MTBE)加醋酸铵对肝脏组织最有效(707 个脂质,变异系数为 30%),BUME(1:1)对心脏组织最有效(311 个脂质,变异系数为 30%)。这些研究结果表明,最有效的脂质提取方法具有高度的组织特异性,强调了针对每种组织类型定制方案的迫切需要。通过对 C57BL/6 小鼠进行干预研究,对优化的组织特异性方法进行了验证,以调查饮食引起的代谢变化。结果表明,每种组织类型都有独特的鉴别性脂质特征,高脂饮食(HFD)和正常饮食(ND)脂肪组织之间有显著差异,共有 13 个亚类中的 374 种脂质,而高脂饮食和正常饮食肝脏组织之间有显著差异,共有 17 个亚类中的 485 种脂质。我们的系统评估证明,量身定制的组织特异性提取方案在全面的脂质组学研究中极具价值,它为可靠地识别脂质变化提供了强大的工具,有助于在各种研究和临床应用中更深入地了解组织特异性代谢过程。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Evaluation of Tissue-Specific Extraction Protocols for Comprehensive Lipid Profiling
Background
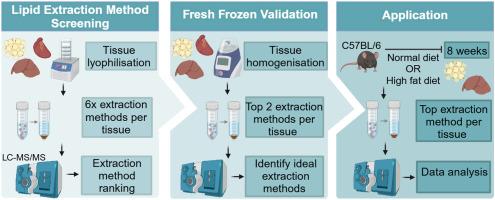
Robust tissue pre-treatment and lipid extraction workflows are crucial to metabolic phenotyping studies and accurate interpretation of lipid profiles. Numerous methods for lipid extraction from tissues have been developed, but the choice of technique influences analysis. This study provides a comprehensive evaluation of six liquid-liquid extraction methods (three biphasic and three monophasic) used for lipidomic tissue analysis by liquid chromatography-mass spectrometry. Extraction methods were assessed for their suitability for comprehensive lipid profiling across diverse tissue types: adipose, liver, and heart. These techniques were compared using lyophilised and fresh frozen samples.Results
The study revealed significant differences in the coverage and reliability of lipid species extracted using each technique, dependent on the tissue type. The optimal extraction method for adipose tissue was butanol:methanol (BUME) (3:1) which achieved the highest lipid coverage, yield and reproducibility (886 lipids with a coefficient of variation (CV) < 30%); methyl tert-butyl ether (MTBE) with ammonium acetate was most effective for liver tissue (707 lipids CV < 30%) and BUME (1:1) for heart tissue (311 lipids CV < 30%). These findings reveal that the most effective lipid extraction methods are highly tissue-specific, underscoring the critical need for bespoke protocols tailored to each tissue type. The optimised tissue-specific methods were validated using an intervention study in C57BL/6 mice to investigate diet-induced metabolic changes. The results demonstrated distinct discriminating lipid profiles unique to each tissue type, with 374 lipid species from 13 subclasses significantly different between high-fat diet (HFD) and normal diet (ND adipose tissue, while 485 lipid species from 17 subclasses were significantly different between HFD and ND liver tissue.Significance and novelty
This study presents a new approach to studying lipid profiles derived from diverse tissues that substantially improve comprehensive lipid species’ detection sensitivity and reliability. Our systematic evaluation provides evidence that tailored tissue-specific extraction protocols are highly valuable in comprehensive lipidomics studies, offering robust tools for reliably identifying lipid changes and facilitates a deeper understanding of tissue-specific metabolic processes in diverse research and clinical applications.求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


