深度学习和单分子定位显微镜揭示DNA纠缠位点的纳米级动力学
IF 16
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
摘要
由于当前成像技术的时间分辨率限制,在纳米尺度上理解分子动力学仍然具有挑战性。与单分子定位显微镜(SMLM)集成的深度学习为探测这些动态提供了机会。在这里,我们利用这种集成来揭示纠缠聚合物在快速时间尺度上的动力学,这在单分子水平上理解相对较少。我们以Lambda DNA为模型系统,利用自回避虫状链模型对它们的纠缠进行建模,沿轮廓生成模拟定位,并在这些模拟图像上训练深度学习算法,从稀疏定位数据中预测链的轮廓。我们发现定位沿等高线呈非均匀分布。我们的评估表明,链缠结为切换缓冲分子创造了局部扩散障碍,影响了在离散DNA片段上与DNA分子共轭的荧光染料的光开关动力学。跟踪这些片段显示了纠缠位点的随机和亚扩散迁移。我们的方法提供了纳米级聚合物动力学和局部分子环境的直接可视化,这是传统成像技术以前无法实现的。此外,我们的研究结果表明,SMLM中荧光团的开关动力学可以用来表征纳米尺度的局部环境。本文章由计算机程序翻译,如有差异,请以英文原文为准。
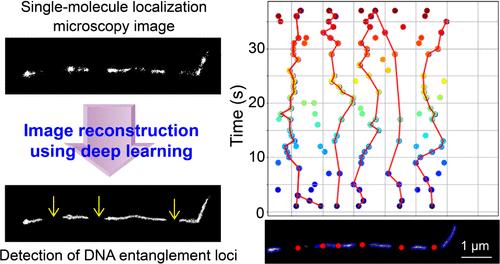
Deep Learning and Single-Molecule Localization Microscopy Reveal Nanoscopic Dynamics of DNA Entanglement Loci
Understanding molecular dynamics at the nanoscale remains challenging due to limitations in the temporal resolution of current imaging techniques. Deep learning integrated with Single-Molecule Localization Microscopy (SMLM) offers opportunities to probe these dynamics. Here, we leverage this integration to reveal entangled polymer dynamics at a fast time scale, which is relatively poorly understood at the single-molecule level. We used Lambda DNA as a model system and modeled their entanglement using the self-avoiding wormlike chain model, generated simulated localizations along the contours, and trained the deep learning algorithm on these simulated images to predict chain contours from sparse localization data. We found that the localizations are heterogeneously distributed along the contours. Our assessments indicated that chain entanglement creates local diffusion barriers for switching buffer molecules, affecting the photoswitching kinetics of fluorescent dyes conjugated to the DNA molecules at discrete DNA segments. Tracking these segments demonstrated stochastic and subdiffusive migration of the entanglement loci. Our approach provides direct visualization of nanoscale polymer dynamics and local molecular environments previously inaccessible to conventional imaging techniques. In addition, our results suggest that the switching kinetics of the fluorophores in SMLM can be used to characterize nanoscopic local environments.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


