早期肠道微生物组中纵向噬菌体-细菌动态
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
人类肠道的微生物定植发生在出生后不久,经过充分研究的阶段,并受到生活方式和其他因素的影响。由于研究规模小,无法利用大型数据库和缺乏适当的生物信息学工具,对婴儿肠道定植过程中噬菌体群落动态知之甚少。作为“青少年糖尿病的环境决定因素”(TEDDY)研究的一部分,我们重新分析了来自四个国家的887名儿童的12262个纵向样本的整个微生物群落霰弹枪测序数据。我们使用Marker-MAGu管道开发了一个广泛的宏基因组组装基因组目录,其中包括来自现有人类微生物组数据集的49111个噬菌体分类群。这被用于鉴定噬菌体标记基因,并将其整合到MetaPhlAn 4细菌标记基因数据库中,从而可以同时评估噬菌体和细菌的动态。我们发现,儿童个体被数百种不同的噬菌体定植,这些噬菌体比细菌更短暂,随着时间的推移,积累了更多样化的噬菌体群落。1型糖尿病与1岁和2岁儿童细菌和病毒群落变化率降低相关。噬菌体数据的添加提高了机器学习模型按国家区分样本的能力。最后,虽然噬菌体种群是特定于个体的,但我们观察到噬菌体生态演替的趋势与假定的宿主细菌密切相关。这一资源提高了我们对早期生命微生物群中噬菌体-细菌相互作用的理解。本文章由计算机程序翻译,如有差异,请以英文原文为准。

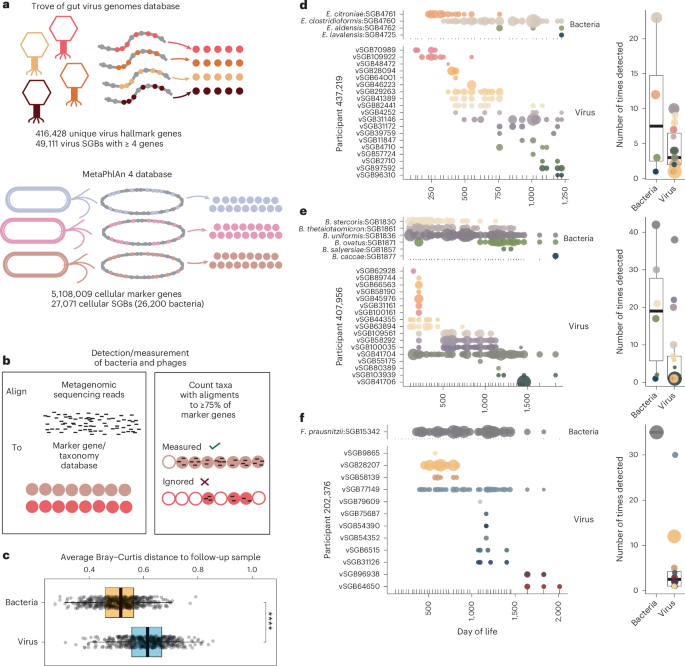
Longitudinal phage–bacteria dynamics in the early life gut microbiome
Microbial colonization of the human gut occurs soon after birth, proceeds through well-studied phases and is affected by lifestyle and other factors. Less is known about phage community dynamics during infant gut colonization due to small study sizes, an inability to leverage large databases and a lack of appropriate bioinformatics tools. Here we reanalysed whole microbial community shotgun sequencing data of 12,262 longitudinal samples from 887 children from four countries across four years of life as part of the The Environmental Determinants of Diabetes in the Young (TEDDY) study. We developed an extensive metagenome-assembled genome catalogue using the Marker-MAGu pipeline, which comprised 49,111 phage taxa from existing human microbiome datasets. This was used to identify phage marker genes and their integration into the MetaPhlAn 4 bacterial marker gene database enabled simultaneous assessment of phage and bacterial dynamics. We found that individual children are colonized by hundreds of different phages, which are more transitory than bacteria, accumulating a more diverse phage community over time. Type 1 diabetes correlated with a decreased rate of change in bacterial and viral communities in children aged one and two. The addition of phage data improved the ability of machine learning models to discriminate samples by country. Finally, although phage populations were specific to individuals, we observed trends of phage ecological succession that correlated well with putative host bacteria. This resource improves our understanding of phage–bacteria interactions in the developing early life microbiome. Reanalysis of 12,262 longitudinal infant gut microbiome samples using the Marker-MAGu pipeline revealed phage–bacteria dynamics over the first year of life.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


