在患者来源的类器官中快速和可扩展的个性化ASO筛查
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
个体化反义寡核苷酸(ASOs)在治疗罕见遗传病方面取得了积极成果。随着临床测序技术的不断进步,识别罕见疾病患者的能力可能会提高,这些患者携带的致病基因变异适合这种治疗策略。在这里,我们描述了一个可扩展的平台,用于生成患者衍生的细胞模型,并证明这些个性化模型可用于患者特异性ASOs的临床前评估。我们描述了将ASO传递到患者来源的类器官模型的方案,并证实了来自杜氏肌营养不良症(DMD)患者的心脏类器官中疾病相关表型的逆转,该患者编码肌营养不良蛋白(DMD)的基因存在结构性缺失,可以用现有的ASO疗法进行治疗。此外,我们为另外两名DMD(兄弟姐妹)患者设计了新的患者特异性aso,这些患者在DMD基因中具有深内含子变异,该变异会产生新的剪接受体位点,结合一个隐外显子和转录产物过早终止。我们发现,用患者特异性ASOs治疗患者源性心脏类器官可恢复DMD表达并逆转疾病相关表型。本文概述的方法为针对多种罕见疾病的个性化ASO疗法的设计和临床前评估提供了快速途径。本文章由计算机程序翻译,如有差异,请以英文原文为准。

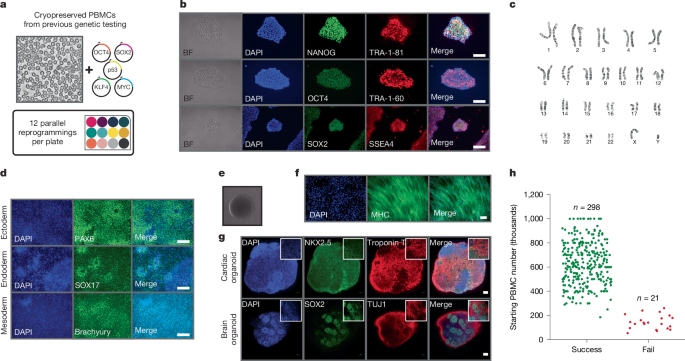
Rapid and scalable personalized ASO screening in patient-derived organoids
Personalized antisense oligonucleotides (ASOs) have achieved positive results in the treatment of rare genetic disease1. As clinical sequencing technologies continue to advance, the ability to identify patients with rare disease harbouring pathogenic genetic variants amenable to this therapeutic strategy will probably improve. Here we describe a scalable platform for generating patient-derived cellular models and demonstrate that these personalized models can be used for preclinical evaluation of patient-specific ASOs. We describe protocols for delivery of ASOs to patient-derived organoid models and confirm reversal of disease-associated phenotypes in cardiac organoids derived from a patient with Duchenne muscular dystrophy (DMD) with a structural deletion in the gene encoding dystrophin (DMD) that is amenable to treatment with existing ASO therapeutics. Furthermore, we designed novel patient-specific ASOs for two additional patients with DMD (siblings) with a deep intronic variant in the DMD gene that gives rise to a novel splice acceptor site, incorporation of a cryptic exon and premature transcript termination. We showed that treatment of patient-derived cardiac organoids with patient-specific ASOs results in restoration of DMD expression and reversal of disease-associated phenotypes. The approach outlined here provides the foundation for an expedited path towards the design and preclinical evaluation of personalized ASO therapeutics for a broad range of rare diseases. A scalable platform for generating patient-specific organoids for testing personalized oligonucleotide therapeutics is described.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


