8,176 个蝙蝠 RNA 病毒元基因组简编揭示了生态驱动因素和循环动态
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
蝙蝠是许多新出现病毒的天然宿主,这些病毒向人类扩散是一个重大风险,但人们对蝙蝠病毒的多样性和生态知之甚少。在这里,我们通过对来自中国52个地点的40个物种的4143只蝙蝠的器官和拭子样本进行亚转录组测序,生成了8,176个RNA病毒元基因组。由此产生的btcn -病毒组数据库将蝙蝠RNA病毒的多样性扩大了3.4倍以上。btcn病毒组中的一些病毒可追溯到哺乳动物、鸟类、节肢动物、软体动物和植物。饮食、感染动态和环境参数(如湿度和森林覆盖率)决定了病毒的分布。与生活在野外的蝙蝠相比,居住在人类住区的蝙蝠携带了更多种病毒,这些病毒也在人类和家畜中传播,包括中国以前未报告的尼帕病毒和洛维病毒。btcn -病毒组为了解蝙蝠病毒的遗传多样性、生态驱动因素和循环动力学提供了重要见解,突出了对人类住区附近蝙蝠进行监测的必要性。本文章由计算机程序翻译,如有差异,请以英文原文为准。

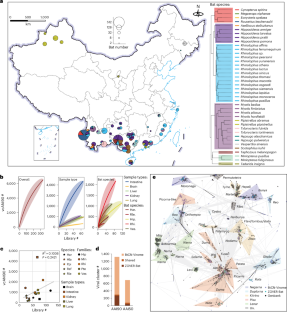
A compendium of 8,176 bat RNA viral metagenomes reveals ecological drivers and circulation dynamics
Bats are natural hosts for many emerging viruses for which spillover to humans is a major risk, but the diversity and ecology of bat viruses is poorly understood. Here we generated 8,176 RNA viral metagenomes by metatranscriptomic sequencing of organ and swab samples from 4,143 bats representing 40 species across 52 locations in China. The resulting database, the BtCN-Virome, expands bat RNA virus diversity by over 3.4-fold. Some viruses in the BtCN-Virome are traced to mammals, birds, arthropods, mollusks and plants. Diet, infection dynamics and environmental parameters such as humidity and forest coverage shape virus distribution. Compared with those in the wild, bats dwelling in human settlements harboured more diverse viruses that also circulated in humans and domestic animals, including Nipah and Lloviu viruses not previously reported in China. The BtCN-Virome provides important insights into the genetic diversity, ecological drivers and circulation dynamics of bat viruses, highlighting the need for surveillance of bats near human settlements. Samples from over 4,000 bats representing 40 different species yield 8,176 viral metagenomes that expand RNA virus diversity and decipher environmental and anthropogenic factors influencing bat viral ecology.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


