淀粉样蛋白原纤维生长的机制Φ-value分析
IF 19.2
1区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
摘要
淀粉样蛋白纤维是高度稳定的错误折叠蛋白质集合体,在多种神经退行性疾病和全身性疾病中发挥着重要作用。尽管目前有关淀粉样蛋白状态的结构信息已经非常丰富,但有关错误折叠过程的机理细节仍然难以捉摸。受蛋白质折叠的Φ值分析的启发,我们结合实验和分子模拟来解析氨基酸接触并确定过渡态组合的结构--PI3K-SH3淀粉样纤维伸长的限速步骤。通过坦福德(Tanford)β分析和自由能计算,实验验证了该组合结构。虽然蛋白质折叠是在漏斗状的能谱上进行的,但在这里我们发现,错误折叠反应的能谱由一个大的 "高尔夫球场 "区域组成,该区域由单一的能障和过渡态定义,并进入一个尖锐的漏斗区域。因此,错误折叠是通过罕见的、成功的单体-纤维末端碰撞发生的,其间夹杂着无数次不成功的结合尝试。综上所述,这些见解为蛋白质错误折叠反应提供了定量和高度解析的描述。本文章由计算机程序翻译,如有差异,请以英文原文为准。

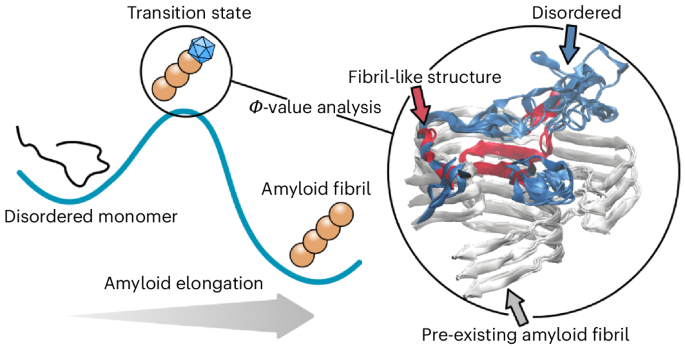
The mechanism of amyloid fibril growth from Φ-value analysis
Amyloid fibrils are highly stable misfolded protein assemblies that play an important role in several neurodegenerative and systemic diseases. Although structural information of the amyloid state is now abundant, mechanistic details about the misfolding process remain elusive. Inspired by the Φ-value analysis of protein folding, we combined experiments and molecular simulations to resolve amino-acid contacts and determine the structure of the transition-state ensemble—the rate-limiting step—for fibril elongation of PI3K-SH3 amyloid fibrils. The ensemble was validated experimentally by Tanford β analysis and computationally by free energy calculations. Although protein folding proceeds on funnel-shaped landscapes, here we find that the energy landscape for the misfolding reaction consists of a large ‘golf course’ region, defined by a single energy barrier and transition state, accessing a sharply funnelled region. Thus, misfolding occurs by rare, successful monomer–fibril end collisions interspersed by numerous unsuccessful binding attempts. Taken together, these insights provide a quantitative and highly resolved description of a protein misfolding reaction. Amyloid fibrils grow through the recruitment of soluble monomer to the fibril end that propagates the fibril structure. Here the transition state, the rate-limiting conformation, of such a reaction has been characterized by Φ-value analysis. An energy landscape model has been developed and fibril growth rates predicted from first principles.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature chemistry
化学-化学综合
CiteScore
29.60
自引率
1.40%
发文量
226
审稿时长
1.7 months
期刊介绍:
Nature Chemistry is a monthly journal that publishes groundbreaking and significant research in all areas of chemistry. It covers traditional subjects such as analytical, inorganic, organic, and physical chemistry, as well as a wide range of other topics including catalysis, computational and theoretical chemistry, and environmental chemistry.
The journal also features interdisciplinary research at the interface of chemistry with biology, materials science, nanotechnology, and physics. Manuscripts detailing such multidisciplinary work are encouraged, as long as the central theme pertains to chemistry.
Aside from primary research, Nature Chemistry publishes review articles, news and views, research highlights from other journals, commentaries, book reviews, correspondence, and analysis of the broader chemical landscape. It also addresses crucial issues related to education, funding, policy, intellectual property, and the societal impact of chemistry.
Nature Chemistry is dedicated to ensuring the highest standards of original research through a fair and rigorous review process. It offers authors maximum visibility for their papers, access to a broad readership, exceptional copy editing and production standards, rapid publication, and independence from academic societies and other vested interests.
Overall, Nature Chemistry aims to be the authoritative voice of the global chemical community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


